iBhb-Lys: Identify lysine β-hydroxybutyrylation sites using autoencoder feature representation and fuzzy SVM algorithm
IF 2.6
4区 生物学
Q2 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
Lysine β-hydroxybutyrylation (Kbhb) is newly discovered β-hydroxybutyrylate-derived histone modification which has been associated with the pathogenesis of many human diseases. To further elucidate the biological significance and molecular mechanism of Kbhb, it is necessary to accurately identify the Kbhb sites from protein sequences. In this study, a novel computational model named iBhb-Lys is developed for the identification of Kbhb sites. Four types of features are combined to encode each Kbhb site as a 3266-dimensional feature vector. And the autoencoder network is used to reduce the dimensionality of feature space, due to the high dimensionality of the combined features. In addition, to effectively reduce the influence of noise and outlier on classification, a new fuzzy support vector machine algorithm is proposed by incorporating the density around the sample into the fuzzy membership function. As illustrated by independent test, the AUC value of iBhb-Lys has increased by 2.22 % compared to the existing predictor KbhbXG. Feature analysis shows that some amino acid composition features, such as the occurrence frequency of leucine and histidine residues around Kbhb sites, contribute profoundly to the identification of Kbhb sites. The conclusions drawn in this study may provide useful reference for studying the molecular mechanism of Kbhb.
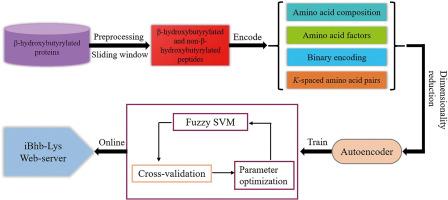
iBhb-Lys:使用自动编码器特征表示和模糊 SVM 算法识别赖氨酸 β-羟基丁酰化位点。
赖氨酸β-羟丁酸化(Kbhb)是新发现的β-羟丁酸衍生组蛋白修饰,它与许多人类疾病的发病机制有关。为了进一步阐明Kbhb的生物学意义和分子机制,有必要从蛋白质序列中准确识别Kbhb位点。本研究开发了一种名为 iBhb-Lys 的新型计算模型,用于识别 Kbhb 位点。该模型结合了四种特征,将每个 Kbhb 位点编码为 3266 维特征向量。由于组合特征的维度较高,因此使用自动编码器网络来降低特征空间的维度。此外,为了有效降低噪声和离群值对分类的影响,还提出了一种新的模糊支持向量机算法,将样本周围的密度纳入模糊成员函数。独立测试表明,iBhb-Lys 的 AUC 值比现有预测器 KbhbXG 提高了 2.22%。特征分析表明,一些氨基酸组成特征,如 Kbhb 位点周围亮氨酸和组氨酸残基的出现频率,对识别 Kbhb 位点有很大帮助。本研究得出的结论可为研究 Kbhb 的分子机理提供有益的参考。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Analytical biochemistry
生物-分析化学
CiteScore
5.70
自引率
0.00%
发文量
283
审稿时长
44 days
期刊介绍:
The journal''s title Analytical Biochemistry: Methods in the Biological Sciences declares its broad scope: methods for the basic biological sciences that include biochemistry, molecular genetics, cell biology, proteomics, immunology, bioinformatics and wherever the frontiers of research take the field.
The emphasis is on methods from the strictly analytical to the more preparative that would include novel approaches to protein purification as well as improvements in cell and organ culture. The actual techniques are equally inclusive ranging from aptamers to zymology.
The journal has been particularly active in:
-Analytical techniques for biological molecules-
Aptamer selection and utilization-
Biosensors-
Chromatography-
Cloning, sequencing and mutagenesis-
Electrochemical methods-
Electrophoresis-
Enzyme characterization methods-
Immunological approaches-
Mass spectrometry of proteins and nucleic acids-
Metabolomics-
Nano level techniques-
Optical spectroscopy in all its forms.
The journal is reluctant to include most drug and strictly clinical studies as there are more suitable publication platforms for these types of papers.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


