Prediction of YY1 loop anchor based on multi-omics features
IF 4.2
3区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
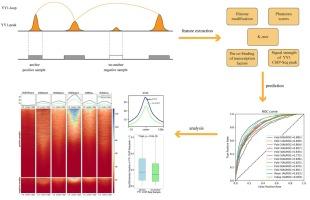
The three-dimensional structure of chromatin is crucial for the regulation of gene expression. YY1 promotes enhancer-promoter interactions in a manner analogous to CTCF-mediated chromatin interactions. However, little is known about which YY1 binding sites can form loop anchors. In this study, the LightGBM model was used to predict YY1-loop anchors by integrating multi-omics data. Due to the large imbalance in the number of positive and negative samples, we use AUPRC to reflect the quality of the classifier. The results show that the LightGBM model exhibits strong predictive performance (). To verify the robustness of the model, the dataset was divided into training and test sets at a 4:1 ratio. The results show that the model performs well for YY1-loop anchor prediction on both the training and independent test sets. Additionally, we ranked the importance of the features and found that the formation of YY1-loop anchors is primarily influenced by the co-binding of transcription factors CTCF, SMC3, and RAD21, as well as histone modifications and sequence context.
基于多组学特征的 YY1 环锚预测
染色质的三维结构对基因表达的调控至关重要。YY1 促进增强子-启动子相互作用的方式类似于 CTCF 介导的染色质相互作用。然而,人们对哪些 YY1 结合位点可以形成环锚知之甚少。本研究利用 LightGBM 模型通过整合多组学数据来预测 YY1 环锚。由于阳性样本和阴性样本的数量存在较大的不平衡,我们使用 AUPRC 来反映分类器的质量。结果表明,LightGBM 模型具有很强的预测性能(AUPRC≥0.93)。为了验证模型的鲁棒性,数据集以 4:1 的比例分为训练集和测试集。结果表明,该模型在训练集和独立测试集上的 YY1 环锚预测性能都很好。此外,我们还对特征的重要性进行了排序,发现 YY1 环锚的形成主要受转录因子 CTCF、SMC3 和 RAD21 的共同结合以及组蛋白修饰和序列上下文的影响。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Methods
生物-生化研究方法
CiteScore
9.80
自引率
2.10%
发文量
222
审稿时长
11.3 weeks
期刊介绍:
Methods focuses on rapidly developing techniques in the experimental biological and medical sciences.
Each topical issue, organized by a guest editor who is an expert in the area covered, consists solely of invited quality articles by specialist authors, many of them reviews. Issues are devoted to specific technical approaches with emphasis on clear detailed descriptions of protocols that allow them to be reproduced easily. The background information provided enables researchers to understand the principles underlying the methods; other helpful sections include comparisons of alternative methods giving the advantages and disadvantages of particular methods, guidance on avoiding potential pitfalls, and suggestions for troubleshooting.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


