Fecal microbial load is a major determinant of gut microbiome variation and a confounder for disease associations
IF 45.5
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
The microbiota in individual habitats differ in both relative composition and absolute abundance. While sequencing approaches determine the relative abundances of taxa and genes, they do not provide information on their absolute abundances. Here, we developed a machine-learning approach to predict fecal microbial loads (microbial cells per gram) solely from relative abundance data. Applying our prediction model to a large-scale metagenomic dataset (n = 34,539), we demonstrated that microbial load is the major determinant of gut microbiome variation and is associated with numerous host factors, including age, diet, and medication. We further found that for several diseases, changes in microbial load, rather than the disease condition itself, more strongly explained alterations in patients’ gut microbiome. Adjusting for this effect substantially reduced the statistical significance of the majority of disease-associated species. Our analysis reveals that the fecal microbial load is a major confounder in microbiome studies, highlighting its importance for understanding microbiome variation in health and disease.
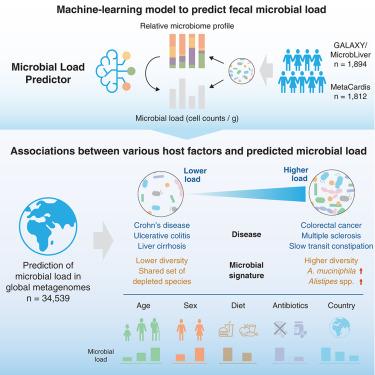
粪便微生物负荷是肠道微生物群变异的主要决定因素,也是疾病关联的混杂因素之一
不同栖息地的微生物群在相对组成和绝对丰度方面都存在差异。虽然测序方法能确定类群和基因的相对丰度,但不能提供其绝对丰度的信息。在这里,我们开发了一种机器学习方法,仅从相对丰度数据预测粪便微生物负荷(每克微生物细胞)。将我们的预测模型应用于大规模元基因组数据集(n = 34,539),我们证明了微生物负荷是肠道微生物组变异的主要决定因素,并且与多种宿主因素有关,包括年龄、饮食和药物。我们进一步发现,对于几种疾病来说,微生物负荷的变化,而不是疾病本身,更能解释患者肠道微生物组的变化。对这一影响进行调整后,大多数疾病相关物种的统计学意义大大降低。我们的分析表明,粪便微生物负荷是微生物组研究中的一个主要混淆因素,突出了其对理解健康和疾病中微生物组变异的重要性。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Cell
生物-生化与分子生物学
CiteScore
110.00
自引率
0.80%
发文量
396
审稿时长
2 months
期刊介绍:
Cells is an international, peer-reviewed, open access journal that focuses on cell biology, molecular biology, and biophysics. It is affiliated with several societies, including the Spanish Society for Biochemistry and Molecular Biology (SEBBM), Nordic Autophagy Society (NAS), Spanish Society of Hematology and Hemotherapy (SEHH), and Society for Regenerative Medicine (Russian Federation) (RPO).
The journal publishes research findings of significant importance in various areas of experimental biology, such as cell biology, molecular biology, neuroscience, immunology, virology, microbiology, cancer, human genetics, systems biology, signaling, and disease mechanisms and therapeutics. The primary criterion for considering papers is whether the results contribute to significant conceptual advances or raise thought-provoking questions and hypotheses related to interesting and important biological inquiries.
In addition to primary research articles presented in four formats, Cells also features review and opinion articles in its "leading edge" section, discussing recent research advancements and topics of interest to its wide readership.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


