Expanding the diversity of origin of transfer-containing sequences in mobilizable plasmids
IF 20.5
1区 生物学
Q1 MICROBIOLOGY
引用次数: 0
Abstract
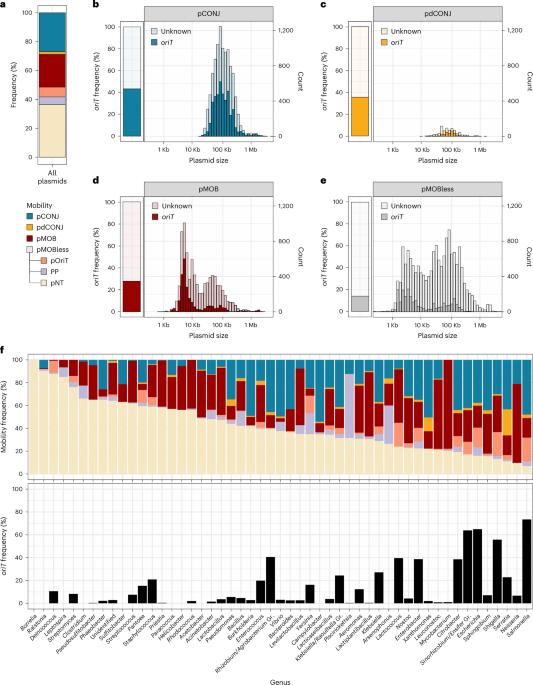
Conjugative plasmids are important drivers of bacterial evolution. Most plasmids lack genes for conjugation and characterized origins of transfer (oriT), which has hampered our understanding of plasmid mobility. Here we used bioinformatic analyses to characterize occurrences of known oriT families across 38,057 plasmids, confirming that most conjugative and mobilizable plasmids lack identifiable oriTs. Recognizable oriT sequences tend to be intergenic, upstream of relaxase genes and specifically associated with relaxase types. We used these criteria to develop a computational method to search for and identify 21 additional families of oriT-containing sequences in plasmids from the pathogens Escherichia coli, Klebsiella pneumoniae and Acinetobacter baumannii. Sequence analyses found 3,072 occurrences of these oriT-containing sequences across 2,976 plasmids, many of which encoded antimicrobial resistance genes. Six candidate oriT-containing sequences were validated experimentally and were shown to facilitate conjugation in E. coli. These findings expand our understanding of plasmid mobility. Characterization of known plasmid oriT features facilitates identification of 21 oriT-containing sequence families which, alongside candidate sequence validation, expands our understanding of plasmid mobility, including plasmids encoding antibiotic resistance.
扩大可移动质粒中含转移序列起源的多样性
共轭质粒是细菌进化的重要驱动力。大多数质粒缺乏共轭基因和特征性转运起源(oriT),这阻碍了我们对质粒流动性的了解。在这里,我们利用生物信息学分析鉴定了 38057 个质粒中已知 oriT 家族的出现情况,证实大多数共轭和可迁移质粒缺乏可识别的 oriT。可识别的 oriT 序列往往位于基因间、弛豫酶基因的上游,并与弛豫酶类型特别相关。我们利用这些标准开发了一种计算方法,在病原体大肠埃希菌、肺炎克雷伯氏菌和鲍曼不动杆菌的质粒中搜索并鉴定了另外 21 个含 oriT 的序列家族。序列分析发现,这些含 oriT 的序列在 2,976 个质粒中出现了 3,072 次,其中许多编码抗菌药耐药性基因。实验验证了六个候选的含 oriT 序列,结果表明它们能促进大肠杆菌中的共轭作用。这些发现拓展了我们对质粒流动性的理解。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Microbiology
Immunology and Microbiology-Microbiology
CiteScore
44.40
自引率
1.10%
发文量
226
期刊介绍:
Nature Microbiology aims to cover a comprehensive range of topics related to microorganisms. This includes:
Evolution: The journal is interested in exploring the evolutionary aspects of microorganisms. This may include research on their genetic diversity, adaptation, and speciation over time.
Physiology and cell biology: Nature Microbiology seeks to understand the functions and characteristics of microorganisms at the cellular and physiological levels. This may involve studying their metabolism, growth patterns, and cellular processes.
Interactions: The journal focuses on the interactions microorganisms have with each other, as well as their interactions with hosts or the environment. This encompasses investigations into microbial communities, symbiotic relationships, and microbial responses to different environments.
Societal significance: Nature Microbiology recognizes the societal impact of microorganisms and welcomes studies that explore their practical applications. This may include research on microbial diseases, biotechnology, or environmental remediation.
In summary, Nature Microbiology is interested in research related to the evolution, physiology and cell biology of microorganisms, their interactions, and their societal relevance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


