Ab initio characterization of protein molecular dynamics with AI2BMD
IF 50.5
1区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
Abstract
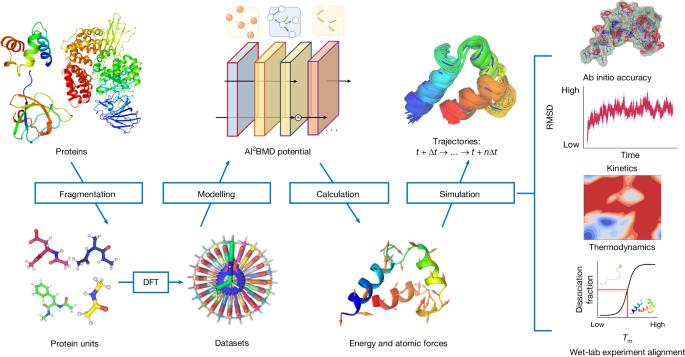
Biomolecular dynamics simulation is a fundamental technology for life sciences research, and its usefulness depends on its accuracy and efficiency1–3. Classical molecular dynamics simulation is fast but lacks chemical accuracy4,5. Quantum chemistry methods such as density functional theory can reach chemical accuracy but cannot scale to support large biomolecules6. Here we introduce an artificial intelligence-based ab initio biomolecular dynamics system (AI2BMD) that can efficiently simulate full-atom large biomolecules with ab initio accuracy. AI2BMD uses a protein fragmentation scheme and a machine learning force field7 to achieve generalizable ab initio accuracy for energy and force calculations for various proteins comprising more than 10,000 atoms. Compared to density functional theory, it reduces the computational time by several orders of magnitude. With several hundred nanoseconds of dynamics simulations, AI2BMD demonstrated its ability to efficiently explore the conformational space of peptides and proteins, deriving accurate 3J couplings that match nuclear magnetic resonance experiments, and showing protein folding and unfolding processes. Furthermore, AI2BMD enables precise free-energy calculations for protein folding, and the estimated thermodynamic properties are well aligned with experiments. AI2BMD could potentially complement wet-lab experiments, detect the dynamic processes of bioactivities and enable biomedical research that is impossible to conduct at present. A study introduces AI2BMD, an artificial intelligence-based dynamics simulation program that uses protein fragmentation with a machine learning force field to accurately and efficiently model the folding and unfolding of large proteins.

利用 AI2BMD 对蛋白质分子动力学进行 Ab initio 表征
生物分子动力学模拟是生命科学研究的基础技术,其实用性取决于其准确性和效率1,2,3。经典分子动力学模拟速度快,但缺乏化学精度4,5。量子化学方法(如密度泛函理论)可以达到化学精度,但无法扩展到支持大型生物分子6。在此,我们介绍一种基于人工智能的ab initio生物分子动力学系统(AI2BMD),它可以高效地模拟具有ab initio精度的全原子大型生物分子。AI2BMD 采用蛋白质破碎方案和机器学习力场7 ,对超过 10,000 个原子的各种蛋白质进行能量和力计算,从而达到可推广的 ab initio 精确度。与密度泛函理论相比,它将计算时间缩短了几个数量级。通过几百纳秒的动力学模拟,AI2BMD 展示了其高效探索肽和蛋白质构象空间的能力,得出了与核磁共振实验相匹配的精确 3J 耦合,并展示了蛋白质的折叠和展开过程。此外,AI2BMD 还能对蛋白质折叠进行精确的自由能计算,估计的热力学性质与实验结果非常吻合。AI2BMD 有可能补充湿实验室实验,检测生物活性的动态过程,并实现目前无法进行的生物医学研究。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature
综合性期刊-综合性期刊
CiteScore
90.00
自引率
1.20%
发文量
3652
审稿时长
3 months
期刊介绍:
Nature is a prestigious international journal that publishes peer-reviewed research in various scientific and technological fields. The selection of articles is based on criteria such as originality, importance, interdisciplinary relevance, timeliness, accessibility, elegance, and surprising conclusions. In addition to showcasing significant scientific advances, Nature delivers rapid, authoritative, insightful news, and interpretation of current and upcoming trends impacting science, scientists, and the broader public. The journal serves a dual purpose: firstly, to promptly share noteworthy scientific advances and foster discussions among scientists, and secondly, to ensure the swift dissemination of scientific results globally, emphasizing their significance for knowledge, culture, and daily life.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


