Probing Single-Molecule Dynamics in Self-Assembling Viral Nucleocapsids
IF 9.6
1区 材料科学
Q1 CHEMISTRY, MULTIDISCIPLINARY
引用次数: 0
Abstract
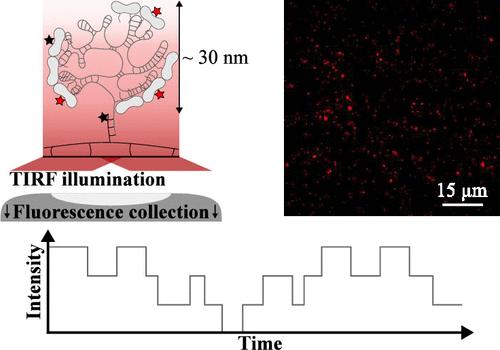
All viruses on Earth rely on host cell machinery for replication, a process that involves a complex self-assembly mechanism. Our aim here is to scrutinize in real time the growth of icosahedral viral nucleocapsids with single-molecule precision. Using total internal reflection fluorescence microscopy, we probed the binding and unbinding dynamics of fluorescently labeled capsid subunits on hundreds of immobilized viral RNA molecules simultaneously at each time point. A step-detection algorithm combined with statistical analysis allowed us to estimate microscopic quantities such as the equilibrium binding rate and mean residence time, which are otherwise inaccessible through traditional ensemble-averaging techniques. Additionally, we could estimate a set of rate constants modeling the growth kinetics from nonequilibrium measurements, and we observed an acceleration in growth caused by the electrostatic screening effect of monovalent salts. Single-molecule fluorescence imaging will be crucial for elucidating virus self-assembly at the molecular level, particularly in crowded, cell-like environments.
自组装病毒核苷酸中的单分子动力学探测
地球上的所有病毒都依赖宿主细胞机制进行复制,这一过程涉及复杂的自组装机制。我们的目的是以单分子精度实时观察二十面体病毒核苷酸的生长过程。利用全内反射荧光显微镜,我们探究了荧光标记的噬菌体亚基在每个时间点上同时与数百个固定的病毒 RNA 分子结合和解除结合的动态。阶跃检测算法与统计分析相结合,使我们能够估算出平衡结合率和平均停留时间等微观量,而传统的集合平均技术是无法估算出这些微观量的。此外,我们还能从非平衡测量中估算出一组模拟生长动力学的速率常数,并观察到单价盐的静电屏蔽效应导致的生长加速。单分子荧光成像对于阐明病毒在分子水平上的自组装至关重要,尤其是在拥挤的类细胞环境中。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nano Letters
工程技术-材料科学:综合
CiteScore
16.80
自引率
2.80%
发文量
1182
审稿时长
1.4 months
期刊介绍:
Nano Letters serves as a dynamic platform for promptly disseminating original results in fundamental, applied, and emerging research across all facets of nanoscience and nanotechnology. A pivotal criterion for inclusion within Nano Letters is the convergence of at least two different areas or disciplines, ensuring a rich interdisciplinary scope. The journal is dedicated to fostering exploration in diverse areas, including:
- Experimental and theoretical findings on physical, chemical, and biological phenomena at the nanoscale
- Synthesis, characterization, and processing of organic, inorganic, polymer, and hybrid nanomaterials through physical, chemical, and biological methodologies
- Modeling and simulation of synthetic, assembly, and interaction processes
- Realization of integrated nanostructures and nano-engineered devices exhibiting advanced performance
- Applications of nanoscale materials in living and environmental systems
Nano Letters is committed to advancing and showcasing groundbreaking research that intersects various domains, fostering innovation and collaboration in the ever-evolving field of nanoscience and nanotechnology.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


