Structural variation reshapes population gene expression and trait variation in 2,105 Brassica napus accessions
IF 31.7
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
Abstract
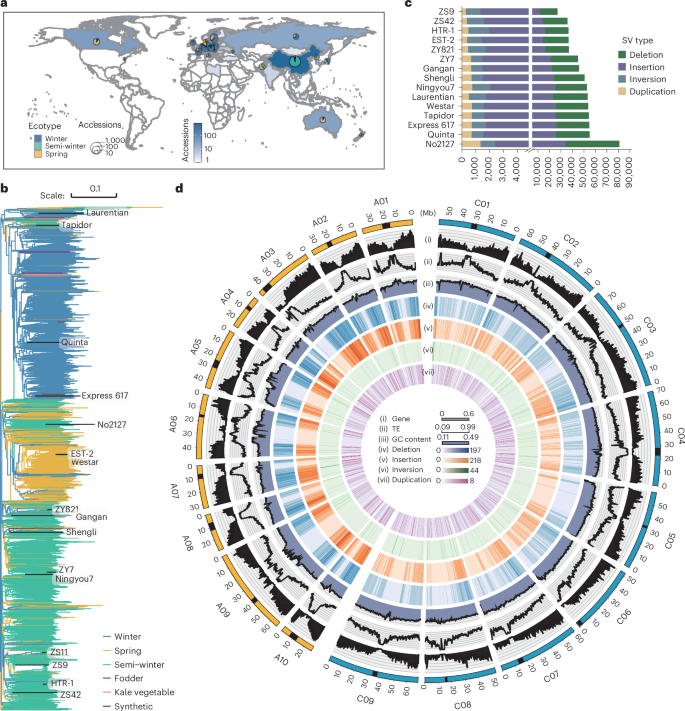
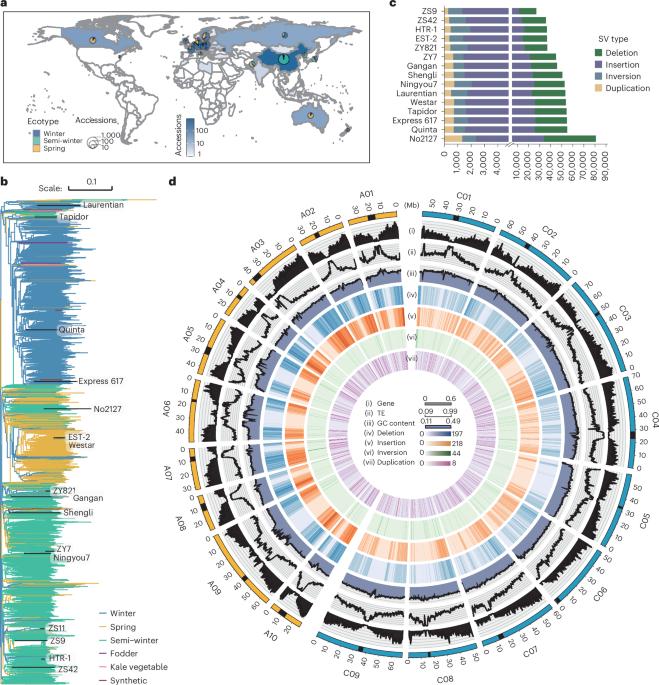
Although individual genomic structural variants (SVs) are known to influence gene expression and trait variation, the extent and scale of SV impact across a species remain unknown. In the present study, we constructed a reference library of 334,461 SVs from genome assemblies of 16 representative morphotypes of neopolyploid Brassica napus accessions and detected 258,865 SVs in 2,105 resequenced genomes. Coupling with 5 tissue population transcriptomes, we uncovered 285,976 SV-expression quantitative trait loci (eQTLs) that associate with altered expression of 73,580 genes. We developed a pipeline for the high-throughput joint analyses of SV-genome-wide association studies (SV-GWASs) and transcriptome-wide association studies of phenomic data, eQTLs and eQTL-GWAS colocalization, and identified 726 SV–gene expression–trait variation associations, some of which were verified by transgenics. The pervasive SV impact on how SV reshapes trait variation was demonstrated with the glucosinolate biosynthesis and transport pathway. The study highlighting the impact of genome-wide and species-scale SVs provides a powerful methodological strategy and valuable resources for studying evolution, gene discovery and breeding. Multiomics joint analyses based on a structural variant (SV) map from 16 genome assemblies and 2,105 resequenced accession genomes shed light on the regulatory effect of SVs on gene expression and trait variation in Brassica napus.
结构变异重塑了 2 105 个油菜品种的群体基因表达和性状变异
尽管已知单个基因组结构变异(SVs)会影响基因表达和性状变异,但 SV 对整个物种的影响程度和规模仍然未知。在本研究中,我们从 16 个具有代表性的新多倍体芸苔属品种的基因组组装中构建了一个包含 334,461 个 SV 的参考文献库,并在 2,105 个重新测序的基因组中检测到 258,865 个 SV。结合 5 个组织群体转录组,我们发现了 285,976 个 SV 表达量性状位点(eQTLs),这些位点与 73,580 个基因的表达改变有关。我们开发了一个高通量联合分析 SV 基因组关联研究(SV-GWAS)和转录组关联研究的表型组数据、eQTL 和 eQTL-GWAS 共定位的管道,并确定了 726 个 SV 基因表达与性状变异的关联,其中一些关联通过转基因得到了验证。葡萄苷酸生物合成和转运途径证明了 SV 对 SV 如何重塑性状变异的普遍影响。这项研究强调了全基因组和物种尺度 SV 的影响,为研究进化、基因发现和育种提供了有力的方法策略和宝贵的资源。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature genetics
生物-遗传学
CiteScore
43.00
自引率
2.60%
发文量
241
审稿时长
3 months
期刊介绍:
Nature Genetics publishes the very highest quality research in genetics. It encompasses genetic and functional genomic studies on human and plant traits and on other model organisms. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation.
Integrative genetic topics comprise, but are not limited to:
-Genes in the pathology of human disease
-Molecular analysis of simple and complex genetic traits
-Cancer genetics
-Agricultural genomics
-Developmental genetics
-Regulatory variation in gene expression
-Strategies and technologies for extracting function from genomic data
-Pharmacological genomics
-Genome evolution
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


