The evolutionary landscape of prokaryotic chromosome/plasmid balance
IF 5.2
1区 生物学
Q1 BIOLOGY
引用次数: 0
Abstract
The balance between chromosomal and plasmid DNAs determines the genomic plasticity of prokaryotes. Natural selections, acting on the level of organisms or plasmids, shape the abundances of plasmid DNAs in prokaryotic genomes. Despite the importance of plasmids in health and engineering, there have been rare systematic attempts to quantitatively model and predict the determinants underlying the strength of different selection forces. Here, we develop a metabolic flux model that describes the intracellular resource competition between chromosomal and plasmid-encoded reactions. By coarse graining, this model predicts a landscape of natural selections on chromosome/plasmid balance, which is featured by the tradeoff between phenotypic and non-phenotypic selection pressures. This landscape is further validated by the observed pattern of plasmid distributions in the vast collection of prokaryotic genomes retrieved from the NCBI database. Our results establish a universal paradigm to understand the prokaryotic chromosome/plasmid interplay and provide insights into the evolutionary origin of plasmid diversity. An intracellular resource competition model sheds light on the evolutionary forces that govern the balance between chromosomal and plasmid DNAs in prokaryotic genomes.
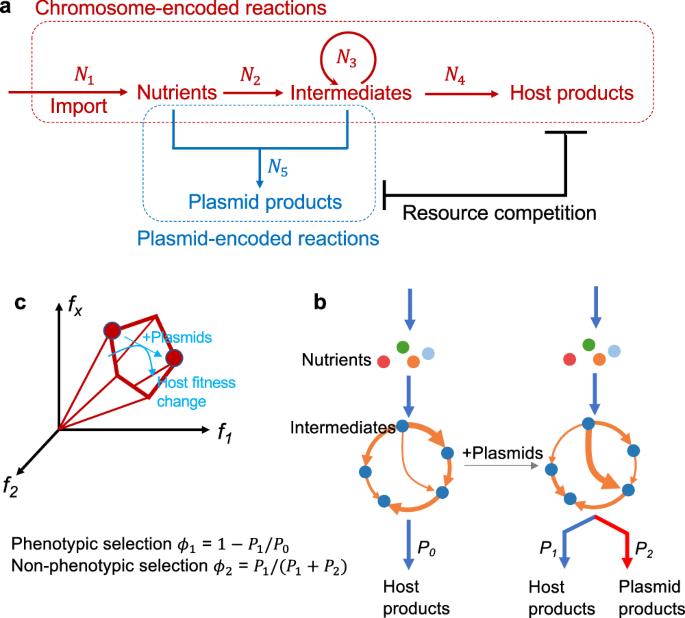
原核生物染色体/质粒平衡的进化图景。
染色体和质粒 DNA 之间的平衡决定了原核生物基因组的可塑性。作用于生物体或质粒层面的自然选择决定了原核生物基因组中质粒 DNA 的丰度。尽管质粒在健康和工程学中非常重要,但很少有人系统地尝试对不同选择力强度的决定因素进行定量建模和预测。在这里,我们建立了一个代谢通量模型,用于描述染色体和质粒编码反应之间的细胞内资源竞争。通过粗粒度分析,该模型预测了染色体/质粒平衡的自然选择景观,其特点是表型和非表型选择压力之间的权衡。从 NCBI 数据库检索到的大量原核生物基因组中观察到的质粒分布模式进一步验证了这一格局。我们的研究结果为理解原核生物染色体/质粒之间的相互作用建立了一个通用范式,并为质粒多样性的进化起源提供了见解。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Communications Biology
Medicine-Medicine (miscellaneous)
CiteScore
8.60
自引率
1.70%
发文量
1233
审稿时长
13 weeks
期刊介绍:
Communications Biology is an open access journal from Nature Research publishing high-quality research, reviews and commentary in all areas of the biological sciences. Research papers published by the journal represent significant advances bringing new biological insight to a specialized area of research.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


