Bile acid conjugated chitosan nanoparticles promote the proliferation and epithelial-mesenchymal transition of hepatocellular carcinoma by regulating the PI3K/Akt/mTOR pathway
IF 2.4
3区 化学
Q3 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
Bile acids have been known to play significant roles at certain physiological levels in gastrointestinal metabolism. Yet, they are known to be carcinogenic and aid in tumor progression in most cases, although the roles remain uncertain. Hence, we tested the cytotoxic potential of cholic acid (CA) loaded chitosan nanoparticles (CNPs) on Hep3B cells. The physicochemical properties of the CNPs synthesized with CA load (CA-CNPs) were determined using standard techniques such as ultraviolet–visible spectrophotometry (UV–Vis), fourier-transform infrared spectroscopy (FTIR), X-ray diffraction (XRD), dynamic light scattering (DLS) and transmission electron microscopy (TEM). The characteristic peak for chitosan nanoparticles were observed for plain CNPs (pCNPs) and CA-CNPs at around 300 nm as per UV–Vis analysis. FTIR analysis indicated the possible trapping of CA onto CNPs as certain peaks were retained and some peaks were shifted. XRD analysis determined that the peaks representing CA and pCNPs were collectively obtained in CA-CNPs. As per DLS analysis, the particle size, PDI and ζ-potential of the CA-CNPs were 259 nm, 0.284 and 30.4 mV. Further, the CA-CNPs were non-cytotoxic on Hep3B cells at the maximum tested concentration of 500 μg/mL. The viability at 500 μg/mL of CA-CNPs was two-fold higher than 500 μg/mL of pCNPs. Also, the pCNPs were not hemolytic and therefore could not have played a role in the increase of viability after treatment with CA-CNPs, which indicates that CA posed a major role in increased viability of Hep3B cells. As per quantitative PCR (qPCR), the upregulated gene expressions of PI3K, Akt, mTORC2, cMyc, Fibronectin, hVPS34, Slug and ZEB1 and the downregulated expression of the tumor suppressor PTEN indicates that PI3K/Akt/mTOR pathway mediated the induction of epithelial-to-mesenchymal transition (EMT) in response to CA-CNPs treatment on Hep3B cells.
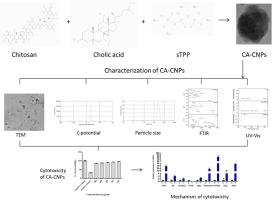
胆汁酸共轭壳聚糖纳米粒子通过调节 PI3K/Akt/mTOR 通路促进肝细胞癌的增殖和上皮-间质转化。
众所周知,胆汁酸在胃肠道代谢的某些生理水平上发挥着重要作用。然而,众所周知,胆汁酸在大多数情况下具有致癌性并有助于肿瘤进展,尽管其作用仍不确定。因此,我们测试了胆酸(CA)负载壳聚糖纳米颗粒(CNPs)对 Hep3B 细胞的细胞毒性潜力。我们采用紫外-可见分光光度法(UV-Vis)、傅立叶变换红外光谱法(FTIR)、X射线衍射法(XRD)、动态光散射法(DLS)和透射电子显微镜法(TEM)等标准技术测定了载CA的壳聚糖纳米颗粒(CNPs)的理化性质。根据紫外可见光分析,普通 CNPs(pCNPs)和 CA-CNPs 的壳聚糖纳米颗粒在 300 纳米左右出现特征峰。傅立叶变换红外分析表明,CA 可能被捕获到 CNP 上,因为某些峰值被保留下来,而某些峰值发生了偏移。XRD 分析表明,在 CA-CNPs 中,代表 CA 和 pCNPs 的峰是共同的。根据 DLS 分析,CA-CNPs 的粒度、PDI 和 ζ 电位分别为 259 nm、0.284 和 30.4 mV。此外,在最大测试浓度为 500 μg/mL 时,CA-CNPs 对 Hep3B 细胞无细胞毒性。浓度为 500 μg/mL 的 CA-CNPs 的存活率是浓度为 500 μg/mL 的 pCNPs 的两倍。此外,pCNPs 不溶血,因此不可能对 CA-CNPs 处理后的活力增加起作用,这表明 CA 对 Hep3B 细胞活力的增加起了主要作用。根据定量 PCR(qPCR),PI3K、Akt、mTORC2、cMyc、Fibronectin、hVPS34、Slug 和 ZEB1 的基因表达上调,而肿瘤抑制因子 PTEN 的表达下调,这表明 CA-CNPs 处理 Hep3B 细胞后,PI3K/Akt/mTOR 通路介导了上皮细胞向间质转化(EMT)的诱导。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Carbohydrate Research
化学-生化与分子生物学
CiteScore
5.00
自引率
3.20%
发文量
183
审稿时长
3.6 weeks
期刊介绍:
Carbohydrate Research publishes reports of original research in the following areas of carbohydrate science: action of enzymes, analytical chemistry, biochemistry (biosynthesis, degradation, structural and functional biochemistry, conformation, molecular recognition, enzyme mechanisms, carbohydrate-processing enzymes, including glycosidases and glycosyltransferases), chemical synthesis, isolation of natural products, physicochemical studies, reactions and their mechanisms, the study of structures and stereochemistry, and technological aspects.
Papers on polysaccharides should have a "molecular" component; that is a paper on new or modified polysaccharides should include structural information and characterization in addition to the usual studies of rheological properties and the like. A paper on a new, naturally occurring polysaccharide should include structural information, defining monosaccharide components and linkage sequence.
Papers devoted wholly or partly to X-ray crystallographic studies, or to computational aspects (molecular mechanics or molecular orbital calculations, simulations via molecular dynamics), will be considered if they meet certain criteria. For computational papers the requirements are that the methods used be specified in sufficient detail to permit replication of the results, and that the conclusions be shown to have relevance to experimental observations - the authors'' own data or data from the literature. Specific directions for the presentation of X-ray data are given below under Results and "discussion".
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


