Polyclonal-to-monoclonal transition in colorectal precancerous evolution
IF 48.5
1区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
Abstract
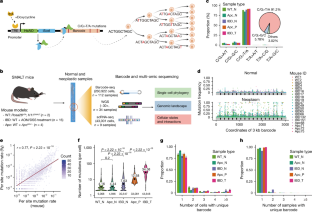
Unravelling the origin and evolution of precancerous lesions is crucial for effectively preventing malignant transformation, yet our current knowledge remains limited1–3. Here we used a base editor-enabled DNA barcoding system4 to comprehensively map single-cell phylogenies in mouse models of intestinal tumorigenesis induced by inflammation or loss of the Apc gene. Through quantitative analysis of high-resolution phylogenies including 260,922 single cells from normal, inflamed and neoplastic intestinal tissues, we identified tens of independent cell lineages undergoing parallel clonal expansions within each lesion. We also found polyclonal origins of human sporadic colorectal polyps through bulk whole-exome sequencing and single-gland whole-genome sequencing. Genomic and clinical data support a model of polyclonal-to-monoclonal transition, with monoclonal lesions representing a more advanced stage. Single-cell RNA sequencing revealed extensive intercellular interactions in early polyclonal lesions, but there was significant loss of interactions during monoclonal transition. Therefore, our data suggest that colorectal precancer is often founded by many different lineages and highlight their cooperative interactions in the earliest stages of cancer formation. These findings provide insights into opportunities for earlier intervention in colorectal cancer. Experiments using DNA barcoding for lineage tracing in mouse models of colorectal cancer reveal polyclonal origins of premalignant lesions, with a transition to monoclonality as they progress to the advanced tumour.

大肠癌癌前病变中的多克隆向单克隆转变
了解癌前病变的起源和演变对于有效预防恶性转化至关重要,但我们目前的知识仍然有限1,2,3。在这里,我们利用碱基编辑器支持的 DNA 条形码系统4,在炎症或 Apc 基因缺失诱导的小鼠肠道肿瘤发生模型中全面绘制了单细胞系统发生图。通过对来自正常、炎症和肿瘤性肠道组织的 260,922 个单细胞的高分辨率系统发生图进行定量分析,我们确定了在每个病变组织中并行克隆扩增的数十个独立细胞系。通过批量全外显子组测序和单腺全基因组测序,我们还发现了人类散发性结直肠息肉的多克隆起源。基因组和临床数据支持多克隆向单克隆转变的模型,单克隆病变代表更晚期的阶段。单细胞 RNA 测序显示,在早期多克隆病变中存在广泛的细胞间相互作用,但在单克隆转变过程中,细胞间相互作用显著减少。因此,我们的数据表明,结直肠癌前病变往往是由许多不同的细胞系形成的,并强调了它们在癌症形成早期阶段的合作性相互作用。这些发现为早期干预结直肠癌提供了机会。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature
综合性期刊-综合性期刊
CiteScore
90.00
自引率
1.20%
发文量
3652
审稿时长
3 months
期刊介绍:
Nature is a prestigious international journal that publishes peer-reviewed research in various scientific and technological fields. The selection of articles is based on criteria such as originality, importance, interdisciplinary relevance, timeliness, accessibility, elegance, and surprising conclusions. In addition to showcasing significant scientific advances, Nature delivers rapid, authoritative, insightful news, and interpretation of current and upcoming trends impacting science, scientists, and the broader public. The journal serves a dual purpose: firstly, to promptly share noteworthy scientific advances and foster discussions among scientists, and secondly, to ensure the swift dissemination of scientific results globally, emphasizing their significance for knowledge, culture, and daily life.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


