Rapid and accurate bacteria identification through deep-learning-based two-dimensional Raman spectroscopy
IF 5.7
2区 化学
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
Abstract
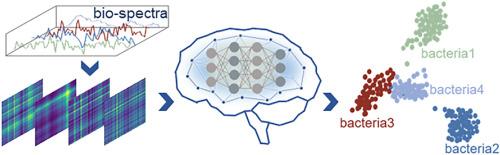
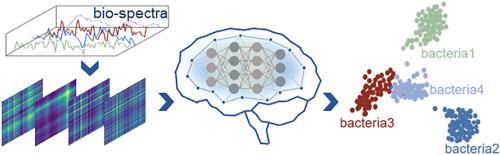
Surface-enhanced Raman spectroscopy (SERS) offers a distinctive vibrational fingerprint of the molecules and has led to widespread applications in medical diagnosis, biochemistry, and virology. With the rapid development of artificial intelligence (AI) technology, AI-enabled Raman spectroscopic techniques, as a promising avenue for biosensing applications, have significantly boosted bacteria identification. By converting spectra into images, the dataset is enriched with more detailed information, allowing AI to identify bacterial isolates with enhanced precision. However, previous studies usually suffer from a trade-off between high-resolution spectrograms for high-accuracy identification and short training time for data processing. Here, we present an efficient bacteria identification strategy that combines deep learning models with a spectrogram encoding algorithm based on wavelet packet transform and Gramian angular field techniques. In contrast to the direct analysis of raw Raman spectra, our approach utilizes wavelet packet transform techniques to compress the spectra by a factor of 1/15, while concurrently maintaining state-of-the-art accuracy by amplifying the subtle differences via Gramian angular field techniques. The results demonstrate that our approach can achieve a 99.64 % and a 90.55 % identification accuracy for two types of bacterial isolates and thirty types of bacterial isolates, respectively, while a 90 % reduction in training time compared to the conventional methods. To verify the model's stability, Gaussian noises were superimposed on the testing dataset, showing a specific generalization ability and superior performance. This algorithm has the potential for integration into on-site testing protocols and is readily updatable with new bacterial isolates. This study provides profound insights and contributes to the current understanding of spectroscopy, paving the way for accurate and rapid bacteria identification in diverse applications of environment monitoring, food safety, microbiology, and public health.
通过基于深度学习的二维拉曼光谱技术快速准确地识别细菌
表面增强拉曼光谱(SERS)提供了分子的独特振动指纹,在医学诊断、生物化学和病毒学领域得到了广泛应用。随着人工智能(AI)技术的快速发展,AI 支持的拉曼光谱技术作为生物传感应用的一个前景广阔的途径,极大地促进了细菌的识别。通过将光谱转换为图像,数据集可获得更多详细信息,从而使人工智能能够更精确地识别细菌分离物。然而,以往的研究通常会在高分辨率光谱图的高精度识别和数据处理的短训练时间之间进行权衡。在这里,我们提出了一种高效的细菌识别策略,它将深度学习模型与基于小波包变换和格拉米安角场技术的频谱图编码算法相结合。与直接分析原始拉曼光谱不同,我们的方法利用小波包变换技术将光谱压缩 1/15 倍,同时通过格拉米安角场技术放大细微差别,从而保持最先进的精度。结果表明,与传统方法相比,我们的方法对两种细菌分离物和 30 种细菌分离物的识别准确率分别达到了 99.64% 和 90.55%,同时减少了 90% 的训练时间。为了验证模型的稳定性,在测试数据集上叠加了高斯噪声,结果表明该模型具有特定的泛化能力和卓越的性能。该算法有望整合到现场测试协议中,并可根据新的细菌分离株随时更新。这项研究提供了深刻的见解,有助于当前对光谱学的理解,为环境监测、食品安全、微生物学和公共卫生等不同应用领域准确、快速地识别细菌铺平了道路。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Analytica Chimica Acta
化学-分析化学
CiteScore
10.40
自引率
6.50%
发文量
1081
审稿时长
38 days
期刊介绍:
Analytica Chimica Acta has an open access mirror journal Analytica Chimica Acta: X, sharing the same aims and scope, editorial team, submission system and rigorous peer review.
Analytica Chimica Acta provides a forum for the rapid publication of original research, and critical, comprehensive reviews dealing with all aspects of fundamental and applied modern analytical chemistry. The journal welcomes the submission of research papers which report studies concerning the development of new and significant analytical methodologies. In determining the suitability of submitted articles for publication, particular scrutiny will be placed on the degree of novelty and impact of the research and the extent to which it adds to the existing body of knowledge in analytical chemistry.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


