Localization of albumin with correlative super resolution light- and electron microscopy in the kidney
IF 5.1
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
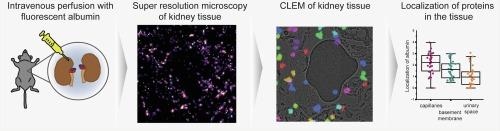
The functioning of vertebrate life relies on renal filtration of surplus fluid and elimination of low-molecular-weight waste products, while keeping serum proteins in the blood. In disease, however, there is leak of serum proteins and tracing them to identify the leaking position within tissue with a nanometer resolution poses a significant challenge. Correlative microscopy integrates the specificity of fluorescent protein labeling into high-resolution electron micrographs. Using chemical tagging of albumin with synthetic fluorophores we achieve protein-specific labeling that preserve their post-embedding fluorescence after high-pressure freezing and freeze-substitution of murine kidney tissue. Using advanced registration techniques for super-resolution correlative light and electron microscopy, we can localize the labeled albumin with a high precision in the x-y plane of electron micrographs and cartograph its distribution. Thereby we can quantify the albumin concentration and measure a linear reduction gradient across the kidney filtration barrier. Our study shows the feasibility of combining different microscopy contrasts for tracing fluorescently labeled protein markers with super resolution in various tissue samples and opens new perspectives for correlative imaging in volume electron microscopy.
利用相关超分辨率光镜和电子显微镜确定肾脏中白蛋白的位置
脊椎动物的生命运作依赖于肾脏过滤多余液体和排出低分子量废物,同时保持血液中的血清蛋白。然而,在疾病情况下,血清蛋白会发生泄漏,要以纳米分辨率追踪血清蛋白以确定其在组织内的泄漏位置,是一项重大挑战。相关显微镜将荧光蛋白标记的特异性与高分辨率电子显微图像相结合。利用合成荧光团对白蛋白进行化学标记,我们实现了蛋白质特异性标记,在对小鼠肾脏组织进行高压冷冻和冷冻置换后仍能保持其包埋后的荧光。利用先进的超分辨率相关光镜和电子显微镜配准技术,我们可以在电子显微图像的 x-y 平面上高精度地定位标记的白蛋白,并绘制其分布图。因此,我们可以量化白蛋白浓度,并测量肾脏滤过屏障上的线性减少梯度。我们的研究表明,结合不同的显微对比度,在各种组织样本中以超分辨率追踪荧光标记的蛋白质标记物是可行的,并为体视电子显微镜的相关成像开辟了新的前景。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Structural Biology: X
Biochemistry, Genetics and Molecular Biology-Structural Biology
CiteScore
6.50
自引率
0.00%
发文量
20
审稿时长
62 days
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


