Autonomous data sampling for high-resolution spatiotemporal fish biomass estimates
IF 5.8
2区 环境科学与生态学
Q1 ECOLOGY
引用次数: 0
Abstract
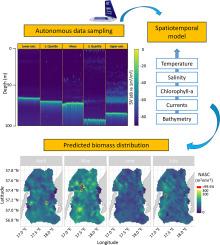
Many key ecological dynamics such as biomass distributions are only detectable on a fine spatiotemporal scale. Autonomous data collection with Unmanned Surface Vehicles (USV) creates new possibilities for cost efficient and high-resolution aquatic data sampling. However, the spatial coverage and sampling resolution remain uncertain due to the novelty of the technology. Further, there is no established method for analysing such fine-scale autocorrelated data without aggregation, potentially compromising data resolution. We here used a USV with an echosounder, a conductivity-temperature sensor and a flourometer to collect data from April–July 2019–2023 in a 60x80km area in the central Baltic Sea. The USV covered a total distance of 8000 nmi, over 42–81 days per year, with an average speed of 0.5 m/s. We combined the hydroacoustic data with publicly available oceanographic data from Copernicus Marine Service Information (CMSI) to describe seasonal distribution dynamics of a small pelagic fish community. Key oceanographic variables collected by the USV were correlated with CMSI estimates at daily/monthly resolution, respectively, to test for suitability to scale (Temperature 0.99/0.97; Salinity −0.77/−0.26; Chlorophyll-a 0.12/0.28). We investigated two approaches of Species Distribution Models (SDMs): generalized additive models (GAM) versus spatiotemporal generalized linear mixed effect models (GLMM). The GLMMs explained the observed data better than the GAMs (R2 0.31 and 0.20, respectively). The addition of environmental variables increased the explanatory capability of GAM and GLMM by 25 % and ∼ 3 %, respectively. Due to the high data resolution, we found significant amounts of positive autocorrelation (R: 0.05–0.30) across more than 50 sequential observations (>6 hours). However, we found that diel patterns in fish detection strongly affected the abundance estimates due to vertically migrating species hiding in the ‘acoustic dead zone’ near the seabed. Such dynamics could only be estimated and corrected for in predictions on the high-resolution data, complicating the trade-off between autocorrelation and high-resolution for SDMs. We compared estimates and effect sizes/directions in identical SDMs on 2x2km/month aggregated (i.e non-autocorrelated) observations and non-aggregated (i.e. autocorrelated) observations, and found relatively little difference in spatiotemporal estimates (r = 0.80). For the first time, we predicted the distribution of a small pelagic fish community at a high spatial resolution, in an area essential to breeding top predators, opening up for new applications in ecological studies locally and globally.
用于高分辨率时空鱼类生物量估算的自主数据采样
许多关键的生态动态(如生物量分布)只能在精细的时空尺度上检测到。利用无人水面航行器(USV)进行自主数据采集,为低成本、高分辨率的水生数据采样创造了新的可能性。然而,由于技术的新颖性,空间覆盖范围和采样分辨率仍不确定。此外,目前还没有一种成熟的方法,可以在不进行汇总的情况下分析这种微尺度自相关数据,从而可能影响数据分辨率。在此,我们使用一艘配备回声测深仪、电导率-温度传感器和浮力计的 USV,从 2019 年 4 月至 7 月,在波罗的海中部 60x80 公里的区域内收集数据。USV 的总航程为 8000 nmi,每年 42-81 天,平均航速为 0.5 m/s。我们将水声数据与哥白尼海洋服务信息(CMSI)提供的公开海洋学数据相结合,描述了小型中上层鱼类群落的季节性分布动态。USV 收集的关键海洋变量分别与哥白尼海洋服务信息(CMSI)的日/月分辨率估计值相关,以检验是否适合比例尺(温度 0.99/0.97;盐度 -0.77/-0.26;叶绿素-a 0.12/0.28)。我们研究了物种分布模型(SDM)的两种方法:广义相加模型(GAM)和时空广义线性混合效应模型(GLMM)。GLMM 比 GAM 更好地解释了观测数据(R2 分别为 0.31 和 0.20)。加入环境变量后,GAM 和 GLMM 的解释能力分别提高了 25% 和 ∼ 3%。由于数据分辨率较高,我们在 50 多个连续观测值(6 小时)中发现了大量正自相关(R:0.05-0.30)。然而,我们发现,由于垂直洄游的鱼类躲藏在海底附近的 "声学死亡区",鱼类探测的昼夜模式严重影响了丰度估算。这种动态变化只能在对高分辨率数据的预测中进行估算和校正,从而使自相关性和高分辨率之间的权衡变得更加复杂。我们比较了 2x2km/month 聚合(即非自相关)观测数据和非聚合(即自相关)观测数据中相同的 SDM 的估计值和效应大小/方向,发现时空估计值的差异相对较小(r = 0.80)。我们首次以较高的空间分辨率预测了小型中上层鱼类群落在顶级掠食者繁殖重要区域的分布,为本地和全球生态研究开辟了新的应用领域。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Ecological Informatics
环境科学-生态学
CiteScore
8.30
自引率
11.80%
发文量
346
审稿时长
46 days
期刊介绍:
The journal Ecological Informatics is devoted to the publication of high quality, peer-reviewed articles on all aspects of computational ecology, data science and biogeography. The scope of the journal takes into account the data-intensive nature of ecology, the growing capacity of information technology to access, harness and leverage complex data as well as the critical need for informing sustainable management in view of global environmental and climate change.
The nature of the journal is interdisciplinary at the crossover between ecology and informatics. It focuses on novel concepts and techniques for image- and genome-based monitoring and interpretation, sensor- and multimedia-based data acquisition, internet-based data archiving and sharing, data assimilation, modelling and prediction of ecological data.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


