Functional study of residual iCre activity relevant for split-Cre applications
IF 4
Q2 BIOTECHNOLOGY & APPLIED MICROBIOLOGY
引用次数: 0
Abstract
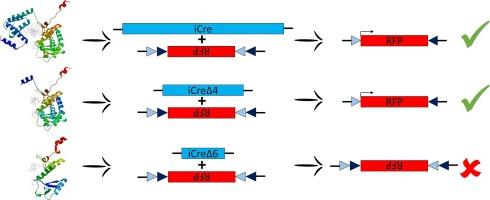
Cre-lox system is a major tool in mouse molecular genetics instrumental in promoting somatic recombination to spatiotemporally control transcriptional activation/inhibition in subsets of cells or tissues in vivo. A critical factor behind this system may be represented by the availability of a specific promoter driving Cre expression in the cell subset of interest. Split-Cre recombinase system represents an evolution that circumvents this limitation using split N- and C-terminal domains of Cre recombinase placed under the control of two distinct promoters defining an intersectional domain where functional complementation of Cre protein fragments is obtained. This system is a valuable tool for controlling Cre recombinase activity in a spatially and temporally defined manner based on the assumption that neither N- or C-terminal Cre fragments alone have recombinase activity. However, residual recombinase activity of one of the two fragments can occur leading to confounding experimental results. In this work, we delve into characterizing functional activity of different N-terminal deleted codon-optimized Cre (iCre) isoforms to refine Split-Cre-based technologies, aiming to avoid uncontrolled recombinase events. Given the presence of several methionine residues in the amino acidic iCre sequence, we explored whether these residues could serve as potential translation start sites, resulting in truncated isoforms that might retain recombinase activity. To address this question, we tested in HEK293T cells whether site-specific recombination was retained in progressively amino-terminal deleted iCre isoforms. Our results reveal residual enzymatic activity of most amino-terminal deleted isoforms of iCre whose ATG start codon is located downstream of the commonly used split site. This insight holds significance for future refinements of the widely used Split-Cre system, providing information to avoid false positive results stemming from unwanted activity.
与分裂-Cre应用相关的残余iCre活性功能研究
Cre-lox 系统是小鼠分子遗传学中的一种重要工具,它有助于促进体细胞重组,从而对体内细胞或组织亚群的转录激活/抑制进行时空控制。该系统背后的一个关键因素可能是在相关细胞亚群中是否存在驱动 Cre 表达的特定启动子。分裂-Cre 重组酶系统代表了一种规避这一限制的进化,它使用分裂的 Cre 重组酶 N 端和 C 端结构域,置于两个不同启动子的控制下,定义了一个交叉结构域,在该结构域中可获得 Cre 蛋白片段的功能互补。该系统是一种宝贵的工具,可用于以空间和时间上明确的方式控制 Cre 重组酶的活性,其假设条件是 N 端或 C 端 Cre 片段单独都不具有重组酶活性。然而,两个片段之一的残余重组酶活性可能会导致实验结果混乱。在这项工作中,我们深入研究了不同的 N 端删除密码子优化 Cre(iCre)异构体的功能活性特征,以完善基于 Split-Cre 的技术,从而避免失控的重组酶事件。鉴于 iCre 氨基酸序列中存在几个蛋氨酸残基,我们探讨了这些残基是否可以作为潜在的翻译起始位点,从而产生可能保留重组酶活性的截短异构体。为了解决这个问题,我们在 HEK293T 细胞中测试了位点特异性重组是否保留在氨基酸末端逐渐缺失的 iCre 异构体中。我们的结果表明,大多数氨基末端缺失的 iCre 异构体(其 ATG 起始密码子位于常用分裂位点的下游)都具有残余的酶活性。这一发现对今后改进广泛使用的 Split-Cre 系统具有重要意义,可为避免因不需要的活性而产生假阳性结果提供信息。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Current Research in Biotechnology
Biochemistry, Genetics and Molecular Biology-Biotechnology
CiteScore
6.70
自引率
3.60%
发文量
50
审稿时长
38 days
期刊介绍:
Current Research in Biotechnology (CRBIOT) is a new primary research, gold open access journal from Elsevier. CRBIOT publishes original papers, reviews, and short communications (including viewpoints and perspectives) resulting from research in biotechnology and biotech-associated disciplines.
Current Research in Biotechnology is a peer-reviewed gold open access (OA) journal and upon acceptance all articles are permanently and freely available. It is a companion to the highly regarded review journal Current Opinion in Biotechnology (2018 CiteScore 8.450) and is part of the Current Opinion and Research (CO+RE) suite of journals. All CO+RE journals leverage the Current Opinion legacy-of editorial excellence, high-impact, and global reach-to ensure they are a widely read resource that is integral to scientists' workflow.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


