Biodegradation of imidacloprid and diuron by Simplicillium sp. QHSH-33
IF 4.2
1区 农林科学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
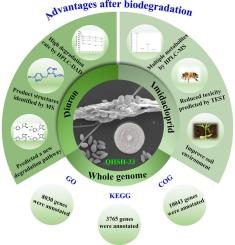
Imidacloprid (IMI) and diuron (DIU) are widely used pesticides in agricultural production. However, their excessive use and high residues have caused harm to the ecological environment and human health. Microbial remediation as an efficient and low-toxic method has become a research hotspot for controlling environmental pollutants. A fungus QHSH-33, identified as Simplicillium sp., has the ability to degrade neonicotinoids IMI and phenylurea DIU. When QHSH-33 and pesticide were co-cultured in liquid medium for 7 days, the degradation rates of IMI and DIU by QHSH-33 in simulated field soil microenvironment were 50.19 % and 70.57 %, respectively. Through HPLC-MS analysis, it was found that the degradation of IMI mainly involved nitro reduction, hydroxylation and other reactions. Three degradation pathways and eight degradation products were identified, among which two metabolites were obtained by microbial transformation of IMI for the first time. The degradation of DIU mainly involved demethylation and dehalogenation reactions, and two degradation pathways and four degradation products were identified, one of which was a new degradation product of DIU. Toxicity assessment demonstrated that most of the degradation products might be considerably less harmful than IMI and DIU. Whole genome sequencing of QHSH-33 revealed a genome size of 33.2 Mbp with 11,707 genes. The genome of QHSH-33 was annotated by KEGG to reveal 128 genes related to exogenous degradation and metabolism. After local blast with reported IMI and DIU degrading enzymes, seven IMI-degrading related genes and seven DIU-degrading related genes were identified in the QHSH-33 genome. The results of this study will help to expand our knowledge on the microbial decomposition metabolism of IMI and DIU, and provide new insights into the degradation mechanism of IMI and DIU in soil and pure culture system, laying a foundation for QHSH-33 strain applied to the removal, biotransformation or detoxification of IMI and DIU.
Simplicillium sp.QHSH-33
吡虫啉(IMI)和杀虫脲(DIU)是农业生产中广泛使用的农药。然而,它们的过量使用和高残留对生态环境和人类健康造成了危害。微生物修复作为一种高效、低毒的方法,已成为控制环境污染物的研究热点。一种被鉴定为 Simplicillium sp.的真菌 QHSH-33 具有降解新烟碱类化合物 IMI 和苯脲类化合物 DIU 的能力。将 QHSH-33 与农药在液体培养基中共同培养 7 天后,在模拟田间土壤微环境中,QHSH-33 对 IMI 和 DIU 的降解率分别为 50.19% 和 70.57%。通过 HPLC-MS 分析发现,IMI 的降解主要涉及硝基还原、羟基化等反应。确定了三种降解途径和八种降解产物,其中有两种代谢物是首次通过微生物转化 IMI 得到的。DIU 的降解主要涉及脱甲基和脱卤反应,确定了两条降解途径和四种降解产物,其中一种是 DIU 的新降解产物。毒性评估表明,大多数降解产物的危害可能大大低于 IMI 和 DIU。QHSH-33 的全基因组测序显示,其基因组大小为 33.2 Mbp,有 11 707 个基因。通过 KEGG 对 QHSH-33 的基因组进行注释,发现了 128 个与外源降解和代谢有关的基因。在对已报道的IMI和DIU降解酶进行局部爆破后,在QHSH-33基因组中发现了7个与IMI降解相关的基因和7个与DIU降解相关的基因。这项研究的结果将有助于拓展我们对IMI和DIU微生物分解代谢的认识,并对IMI和DIU在土壤和纯培养体系中的降解机制提供新的见解,为QHSH-33菌株应用于IMI和DIU的去除、生物转化或解毒奠定基础。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
CiteScore
7.00
自引率
8.50%
发文量
238
审稿时长
4.2 months
期刊介绍:
Pesticide Biochemistry and Physiology publishes original scientific articles pertaining to the mode of action of plant protection agents such as insecticides, fungicides, herbicides, and similar compounds, including nonlethal pest control agents, biosynthesis of pheromones, hormones, and plant resistance agents. Manuscripts may include a biochemical, physiological, or molecular study for an understanding of comparative toxicology or selective toxicity of both target and nontarget organisms. Particular interest will be given to studies on the molecular biology of pest control, toxicology, and pesticide resistance.
Research Areas Emphasized Include the Biochemistry and Physiology of:
• Comparative toxicity
• Mode of action
• Pathophysiology
• Plant growth regulators
• Resistance
• Other effects of pesticides on both parasites and hosts.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


