Multiomic characterization of RNA microenvironments by oligonucleotide-mediated proximity-interactome mapping
IF 36.1
1区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
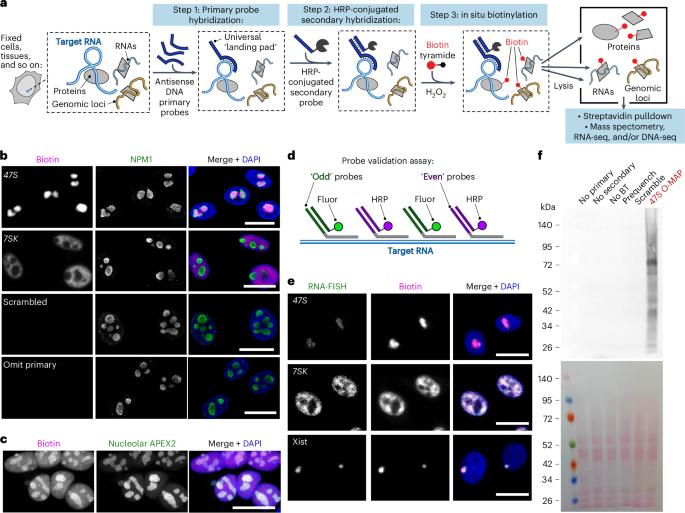
RNA molecules form complex networks of molecular interactions that are central to their function and to cellular architecture. But these interaction networks are difficult to probe in situ. Here, we introduce Oligonucleotide-mediated proximity-interactome MAPping (O-MAP), a method for elucidating the biomolecules near an RNA of interest, within its native context. O-MAP uses RNA-fluorescence in situ hybridization-like oligonucleotide probes to deliver proximity-biotinylating enzymes to a target RNA in situ, enabling nearby molecules to be enriched by streptavidin pulldown. This induces exceptionally precise biotinylation that can be easily optimized and ported to new targets or sample types. Using the noncoding RNAs 47S, 7SK and Xist as models, we develop O-MAP workflows for discovering RNA-proximal proteins, transcripts and genomic loci, yielding a multiomic characterization of these RNAs’ subcellular compartments and new regulatory interactions. O-MAP requires no genetic manipulation, uses exclusively off-the-shelf parts and requires orders of magnitude fewer cells than established methods, making it accessible to most laboratories. Oligonucleotide-mediated proximity-interactome MAPping (O-MAP) enables precise multiomic characterization of biomolecular interaction networks at target RNAs. Distinct O-MAP workflows reveal RNA-adjacent proteins, transcripts and genomic loci.
通过寡核苷酸介导的近端-交互组图谱对 RNA 微环境进行多组表征。
RNA 分子形成复杂的分子相互作用网络,对其功能和细胞结构至关重要。但这些相互作用网络很难进行原位探测。在这里,我们介绍了寡核苷酸介导的邻近相互作用组 MAPping(O-MAP),这是一种在 RNA 的原生环境中阐明其附近生物分子的方法。O-MAP 使用类似于 RNA 荧光原位杂交的寡核苷酸探针,在原位向目标 RNA 运送邻近生物素化酶,从而通过链霉亲和素下拉富集附近的分子。这种方法能诱导异常精确的生物素化,易于优化并可移植到新的靶标或样本类型。以非编码 RNA 47S、7SK 和 Xist 为模型,我们开发了 O-MAP 工作流程,用于发现 RNA 近端蛋白、转录本和基因组位点,从而对这些 RNA 的亚细胞区和新的调控相互作用进行多组表征。O-MAP 不需要基因操作,只使用现成的部件,所需的细胞数量比现有方法少很多,因此大多数实验室都能使用。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Methods
生物-生化研究方法
CiteScore
58.70
自引率
1.70%
发文量
326
审稿时长
1 months
期刊介绍:
Nature Methods is a monthly journal that focuses on publishing innovative methods and substantial enhancements to fundamental life sciences research techniques. Geared towards a diverse, interdisciplinary readership of researchers in academia and industry engaged in laboratory work, the journal offers new tools for research and emphasizes the immediate practical significance of the featured work. It publishes primary research papers and reviews recent technical and methodological advancements, with a particular interest in primary methods papers relevant to the biological and biomedical sciences. This includes methods rooted in chemistry with practical applications for studying biological problems.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


