Deep learning based local feature classification to automatically identify single molecule fluorescence events
IF 5.2
1区 生物学
Q1 BIOLOGY
引用次数: 0
Abstract
Long-term single-molecule fluorescence measurements are widely used powerful tools to study the conformational dynamics of biomolecules in real time to further elucidate their conformational dynamics. Typically, thousands or even more single-molecule traces are analyzed to provide statistically meaningful information, which is labor-intensive and can introduce user bias. Recently, several deep-learning models have been developed to automatically classify single-molecule traces. In this study, we introduce DEBRIS (Deep lEarning Based fRagmentatIon approach for Single-molecule fluorescence event identification), a deep-learning model focusing on classifying local features and capable of automatically identifying steady fluorescence signals and dynamically emerging signals of different patterns. DEBRIS efficiently and accurately identifies both one-color and two-color single-molecule events, including their start and end points. By adjusting user-defined criteria, DEBRIS becomes the pioneer in using a deep learning model to accurately classify four different types of single-molecule fluorescence events using the same trained model, demonstrating its universality and ability to enrich the current toolbox. DEBRIS is a deep learning-based model that can classify local features and identify steady events and dynamically emerging events within two-color single-molecule traces of varying lengths based on user-defined criteria.
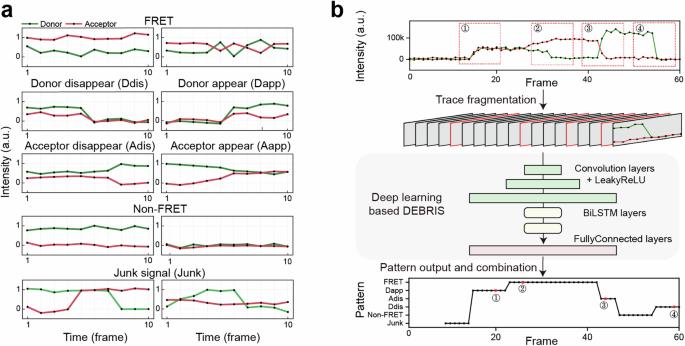
基于深度学习的局部特征分类,自动识别单分子荧光事件。
长期单分子荧光测量是一种广泛使用的强大工具,用于实时研究生物大分子的构象动态,以进一步阐明其构象动态。通常情况下,需要对数千甚至更多的单分子迹线进行分析,才能提供有统计意义的信息,这不仅耗费大量人力,而且可能会引入用户偏差。最近,人们开发了几种深度学习模型来自动对单分子迹线进行分类。在本研究中,我们介绍了 DEBRIS(基于深度学习的单分子荧光事件识别方法),它是一种侧重于局部特征分类的深度学习模型,能够自动识别稳定的荧光信号和不同模式的动态信号。DEBRIS 能高效、准确地识别单色和双色单分子事件,包括其起点和终点。通过调整用户定义的标准,DEBRIS 率先使用深度学习模型,使用同一个训练有素的模型准确地对四种不同类型的单分子荧光事件进行分类,证明了其通用性和丰富当前工具箱的能力。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Communications Biology
Medicine-Medicine (miscellaneous)
CiteScore
8.60
自引率
1.70%
发文量
1233
审稿时长
13 weeks
期刊介绍:
Communications Biology is an open access journal from Nature Research publishing high-quality research, reviews and commentary in all areas of the biological sciences. Research papers published by the journal represent significant advances bringing new biological insight to a specialized area of research.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


