Structural insight into rabies virus neutralization revealed by an engineered antibody scaffold
IF 4.4
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
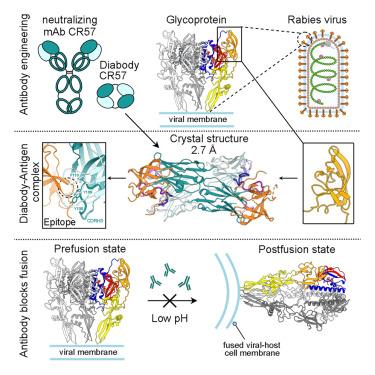
Host-cell entry of the highly pathogenic rabies virus (RABV) is mediated by glycoprotein (G) spikes, which also comprise the primary target for the humoral immune response. RABV glycoprotein (RABV-G) displays several antigenic sites that are targeted by neutralizing monoclonal antibodies (mAbs). In this study, we determined the epitope of a potently neutralizing human mAb, CR57, which we engineered into a diabody format to facilitate crystallization. We report the crystal structure of the CR57 diabody alone at 2.38 Å resolution, and in complex with RABV-G domain III at 2.70 Å resolution. The CR57−RABV-G structure reveals critical interactions at the antigen interface, which target the conserved “KLCGVL” peptide and residues proximal to it on RABV-G. Structural analysis combined with a cell-cell fusion assay demonstrates that CR57 effectively inhibits RABV-G-mediated fusion by obstructing the fusogenic transitions of the spike protein. Altogether, this investigation provides a structural perspective on RABV inhibition by a potently neutralizing human antibody.
工程抗体支架揭示狂犬病病毒中和的结构奥秘
高致病性狂犬病病毒(RABV)通过糖蛋白(G)尖峰进入宿主细胞,这也是体液免疫反应的主要靶点。RABV 糖蛋白(RABV-G)有几个抗原位点,是中和单克隆抗体(mAbs)的靶点。在本研究中,我们确定了一种强效中和人类 mAb CR57 的表位,并将其设计成二抗体形式以方便结晶。我们报告了分辨率为 2.38 Å 的 CR57 二抗体单独晶体结构,以及分辨率为 2.70 Å 的 CR57 与 RABV-G 结构域 III 复合物的晶体结构。CR57-RABV-G 结构揭示了抗原界面上的关键相互作用,其目标是 RABV-G 上保守的 "KLCGVL "肽及其近端残基。结构分析结合细胞-细胞融合试验表明,CR57 通过阻碍尖峰蛋白的融合转换,有效抑制了 RABV-G 介导的融合。总之,这项研究从结构角度揭示了一种强效中和人类抗体对 RABV 的抑制作用。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


