Uracil base PCR implemented for reliable DNA walking
IF 2.6
4区 生物学
Q2 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
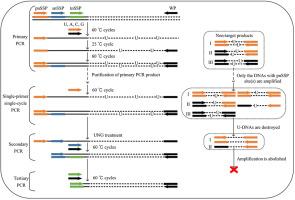
PCR-based DNA walking is of efficacy for capturing unknown flanking genomic sequences. Here, an uracil base PCR (UB-PCR) with satisfying specificity has been devised for DNA walking. Primary UB-PCR replaces thymine base with uracil base, resulting in a primary PCR product composed of U-DNAs. A single-primer (primary nested sequence-specific primer) single-cycle amplification, using the four normal bases (adenine, thymine, cytosine, and guanine) as substrate, is then performed on the primary PCR product. Clearly, only those U-DNAs, ended by the primary nested sequence-specific primer at least at one side, will produce the corresponding normal single strands. Next, the single-cycle product undergoes uracil-DNA glycosylase treatment to destroy the U-DNAs, while the normal single strands are unaffected. Afterward, secondary even tertiary PCR is performed to exclusively enrich the target product. The feasibility of UB-PCR has been checked by obtaining unknown sequences bordering the three selected genetic sites.
实施尿嘧啶碱基 PCR,实现可靠的 DNA 走查。
基于 PCR 的 DNA 走查对于捕获未知的侧翼基因组序列非常有效。在此,我们设计了一种具有良好特异性的尿嘧啶碱基 PCR(UB-PCR)来进行 DNA 扩增。初级 UB-PCR 将胸腺嘧啶碱基替换为尿嘧啶碱基,从而产生由 U-DNA 组成的初级 PCR 产物。然后,以四个正常碱基(腺嘌呤、胸腺嘧啶、胞嘧啶和鸟嘌呤)为底物,对主 PCR 产物进行单引物(主嵌套序列特异性引物)单循环扩增。显然,只有那些至少一边以主嵌套序列特异性引物为末端的 U-DNA 才能产生相应的正常单链。接下来,单循环产物经过尿嘧啶-DNA 糖基化酶处理,以破坏 U-DNA,而正常的单链则不受影响。之后,进行二次甚至三次 PCR,以专门富集目标产物。通过获得与三个选定基因位点相邻的未知序列,检验了 UB-PCR 的可行性。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Analytical biochemistry
生物-分析化学
CiteScore
5.70
自引率
0.00%
发文量
283
审稿时长
44 days
期刊介绍:
The journal''s title Analytical Biochemistry: Methods in the Biological Sciences declares its broad scope: methods for the basic biological sciences that include biochemistry, molecular genetics, cell biology, proteomics, immunology, bioinformatics and wherever the frontiers of research take the field.
The emphasis is on methods from the strictly analytical to the more preparative that would include novel approaches to protein purification as well as improvements in cell and organ culture. The actual techniques are equally inclusive ranging from aptamers to zymology.
The journal has been particularly active in:
-Analytical techniques for biological molecules-
Aptamer selection and utilization-
Biosensors-
Chromatography-
Cloning, sequencing and mutagenesis-
Electrochemical methods-
Electrophoresis-
Enzyme characterization methods-
Immunological approaches-
Mass spectrometry of proteins and nucleic acids-
Metabolomics-
Nano level techniques-
Optical spectroscopy in all its forms.
The journal is reluctant to include most drug and strictly clinical studies as there are more suitable publication platforms for these types of papers.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


