Exploiting cryo-EM structures of actomyosin-5a to reveal the physical properties of its lever
IF 4.4
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
Myosin 5a (Myo5a) is a dimeric processive motor protein that transports cellular cargos along filamentous actin (F-actin). Its long lever is responsible for its large power-stroke, step size, and load-bearing ability. Little is known about the levers’ structure and physical properties, and how they contribute to walking mechanics. Using cryoelectron microscopy (cryo-EM) and molecular dynamics (MD) simulations, we resolved the structure of monomeric Myo5a, comprising the motor domain and full-length lever, bound to F-actin. The range of its lever conformations revealed its physical properties, how stiffness varies along its length and predicts a large, 35 nm, working stroke. Thus, the newly released trail head in a dimeric Myo5a would only need to perform a small diffusive search for its new binding site on F-actin, and stress would only be generated across the dimer once phosphate is released from the lead head, revealing new insight into the walking behavior of Myo5a.
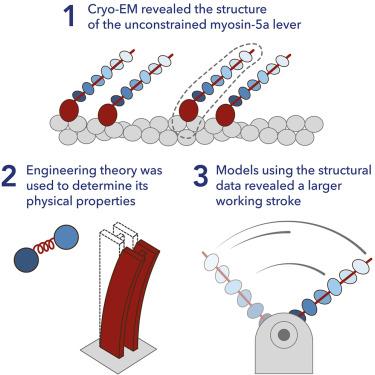
利用肌动蛋白-5a 的低温电子显微镜结构揭示其杠杆的物理特性
肌球蛋白 5a(Myo5a)是一种二聚体过程运动蛋白,可沿着丝状肌动蛋白(F-actin)运输细胞货物。肌球蛋白 5a 的长杠杆使其具有较大的动力冲程、步长和承载能力。人们对杠杆的结构和物理性质以及它们如何对行走机械做出贡献知之甚少。利用冷冻电镜(cryo-EM)和分子动力学(MD)模拟,我们解析了与F-肌动蛋白结合的Myo5a单体结构,包括马达结构域和全长杠杆。其杠杆构象的范围揭示了它的物理特性、刚度如何沿其长度变化,并预测了一个大的、35 nm 的工作冲程。因此,二聚体Myo5a中新释放的踪迹头只需在F-肌动蛋白上对其新的结合位点进行少量的扩散搜索,只有当磷酸从踪迹头释放后,二聚体中才会产生应力,这揭示了Myo5a行走行为的新见解。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


