DNA methylation shapes the Polycomb landscape during the exit from naive pluripotency
IF 12.5
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
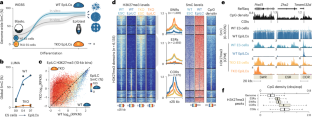
In mammals, 5-methylcytosine (5mC) and Polycomb repressive complex 2 (PRC2)-deposited histone 3 lysine 27 trimethylation (H3K27me3) are generally mutually exclusive at CpG-rich regions. As mouse embryonic stem cells exit the naive pluripotent state, there is massive gain of 5mC concomitantly with restriction of broad H3K27me3 to 5mC-free, CpG-rich regions. To formally assess how 5mC shapes the H3K27me3 landscape, we profiled the epigenome of naive and differentiated cells in the presence and absence of the DNA methylation machinery. Surprisingly, we found that 5mC accumulation is not required to restrict most H3K27me3 domains. Instead, this 5mC-independent H3K27me3 restriction is mediated by aberrant expression of the PRC2 antagonist Ezhip (encoding EZH inhibitory protein). At the subset of regions where 5mC appears to genuinely supplant H3K27me3, we identified 163 candidate genes that appeared to require 5mC deposition and/or H3K27me3 depletion for their activation in differentiated cells. Using site-directed epigenome editing to directly modulate 5mC levels, we demonstrated that 5mC deposition is sufficient to antagonize H3K27me3 deposition and confer gene activation at individual candidates. Altogether, we systematically measured the antagonistic interplay between 5mC and H3K27me3 in a system that recapitulates early embryonic dynamics. Our results suggest that H3K27me3 restraint depends on 5mC, both directly and indirectly. Our study also implies a noncanonical role of 5mC in gene activation, which may be important not only for normal development but also for cancer progression, as oncogenic cells frequently exhibit dynamic replacement of 5mC for H3K27me3 and vice versa. Epigenetic modifications of both DNA and histones undergo profound remodeling during early mammalian development. Epigenome editing demonstrates that DNA methylation can impact certain histone marks, helping control proper gene expression dynamics.

DNA甲基化塑造了摆脱幼稚多能性过程中的多角体景观
在哺乳动物中,5-甲基胞嘧啶(5mC)和多聚胞抑制复合体2(PRC2)沉积的组蛋白3赖氨酸27三甲基化(H3K27me3)通常在富含CpG的区域相互排斥。当小鼠胚胎干细胞脱离幼稚多能状态时,5mC大量增殖,同时宽泛的H3K27me3被限制在不含5mC的富含CpG区域。为了正式评估 5mC 如何影响 H3K27me3 的分布,我们分析了 DNA 甲基化机制存在和不存在时幼稚细胞和分化细胞的表观基因组。令人惊讶的是,我们发现限制大多数 H3K27me3 域并不需要 5mC 的积累。相反,这种不依赖于 5mC 的 H3K27me3 限制是由 PRC2 拮抗剂 Ezhip(编码 EZH 抑制蛋白)的异常表达介导的。在 5mC 似乎真正取代 H3K27me3 的区域子集,我们确定了 163 个候选基因,这些基因似乎需要 5mC 沉积和/或 H3K27me3 消耗才能在分化细胞中激活。利用定点表观遗传组编辑直接调节 5mC 水平,我们证明了 5mC 沉积足以拮抗 H3K27me3 沉积,并赋予单个候选基因活化。总之,我们在一个再现早期胚胎动态的系统中系统地测量了 5mC 和 H3K27me3 之间的拮抗相互作用。我们的研究结果表明,H3K27me3 的抑制作用直接或间接地依赖于 5mC。我们的研究还暗示了 5mC 在基因激活中的非规范作用,这不仅对正常发育很重要,而且对癌症进展也很重要,因为致癌细胞经常表现出 5mC 对 H3K27me3 的动态替代,反之亦然。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Structural & Molecular Biology
BIOCHEMISTRY & MOLECULAR BIOLOGY-BIOPHYSICS
CiteScore
22.00
自引率
1.80%
发文量
160
审稿时长
3-8 weeks
期刊介绍:
Nature Structural & Molecular Biology is a comprehensive platform that combines structural and molecular research. Our journal focuses on exploring the functional and mechanistic aspects of biological processes, emphasizing how molecular components collaborate to achieve a particular function. While structural data can shed light on these insights, our publication does not require them as a prerequisite.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


