Restoring protein glycosylation with GlycoShape
IF 36.1
1区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
Despite ground-breaking innovations in experimental structural biology and protein structure prediction techniques, capturing the structure of the glycans that functionalize proteins remains a challenge. Here we introduce GlycoShape ( https://glycoshape.org ), an open-access glycan structure database and toolbox designed to restore glycoproteins to their native and functional form in seconds. The GlycoShape database counts over 500 unique glycans so far, covering the human glycome and augmented by elements from a wide range of organisms, obtained from 1 ms of cumulative sampling from molecular dynamics simulations. These structures can be linked to proteins with a robust algorithm named Re-Glyco, directly compatible with structural data in open-access repositories, such as the Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB) and AlphaFold Protein Structure Database, or own. The quality, performance and broad applicability of GlycoShape is demonstrated by its ability to predict N-glycosylation occupancy, scoring a 93% agreement with experiment, based on screening all proteins in the PDB with a corresponding glycoproteomics profile, for a total of 4,259 N-glycosylation sequons. GlycoShape is an open-access web-based platform designed to supplement three-dimensional glycoprotein structures with missing structural information on glycans. To link them, the Re-Glyco algorithm evaluates the steric complementarity of glycans using their conformational ensemble with the protein surface.
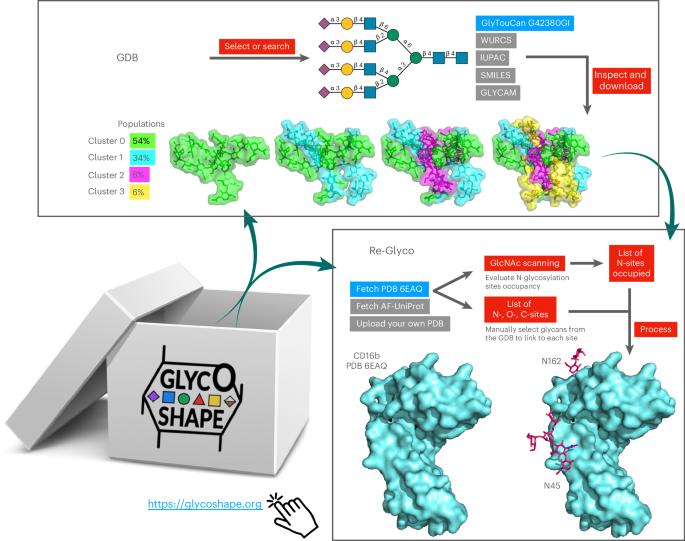
用 GlycoShape 恢复蛋白质糖基化。
尽管在实验结构生物学和蛋白质结构预测技术方面取得了突破性创新,但捕捉使蛋白质功能化的聚糖结构仍然是一项挑战。我们在此介绍 GlycoShape ( https://glycoshape.org ),它是一个开放存取的聚糖结构数据库和工具箱,可在数秒内将糖蛋白还原为原生的功能形式。迄今为止,GlycoShape 数据库已收录了 500 多个独特的聚糖,涵盖了人类聚糖结构,并通过分子动力学模拟 1 毫秒的累积采样获得了多种生物体的聚糖元素。这些结构可以通过一种名为 Re-Glyco 的强大算法与蛋白质连接起来,直接与结构生物信息学研究合作组织蛋白质数据库(RCSB PDB)和 AlphaFold 蛋白结构数据库等开放存取库中的结构数据兼容。GlycoShape 的质量、性能和广泛适用性体现在其预测 N-糖基化占有率的能力上,根据筛选 PDB 中所有蛋白质和相应的糖蛋白组学特征,共 4,259 个 N-糖基化序列,GlycoShape 与实验的吻合度达到 93%。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Methods
生物-生化研究方法
CiteScore
58.70
自引率
1.70%
发文量
326
审稿时长
1 months
期刊介绍:
Nature Methods is a monthly journal that focuses on publishing innovative methods and substantial enhancements to fundamental life sciences research techniques. Geared towards a diverse, interdisciplinary readership of researchers in academia and industry engaged in laboratory work, the journal offers new tools for research and emphasizes the immediate practical significance of the featured work. It publishes primary research papers and reviews recent technical and methodological advancements, with a particular interest in primary methods papers relevant to the biological and biomedical sciences. This includes methods rooted in chemistry with practical applications for studying biological problems.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


