VotePLMs-AFP: Identification of antifreeze proteins using transformer-embedding features and ensemble learning
IF 2.2
3区 生物学
Q3 BIOCHEMISTRY & MOLECULAR BIOLOGY
Biochimica et biophysica acta. General subjects
Pub Date : 2024-10-18
DOI:10.1016/j.bbagen.2024.130721
引用次数: 0
Abstract
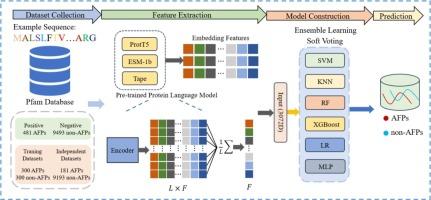
Antifreeze proteins (AFPs) are a unique class of biomolecules capable of protecting other proteins, cell membranes, and cellular structures within organisms from damage caused by freezing conditions. Given the significance of AFPs in various domains such as biotechnology, agriculture, and medicine, several machine learning methods have been developed to identify AFPs. However, due to the complexity and diversity of AFPs, the predictive performance of existing methods is limited. Therefore, there is an urgent need to develop an efficient and rapid computational method for accurately predicting AFPs. In this study, we proposed a novel predictor based on transformer-embedding features and ensemble learning for the identification of AFPs, termed VotePLMs-AFP. Firstly, three types of feature descriptors were extracted from pre-trained protein language models (PLMs) during the feature extraction process. Subsequently, we analyzed six combinations generated by these three embeddings to explore the optimal feature set, which was input into the soft voting-based ensemble learning classifier for the identification of AFPs. Finally, we evaluated the model on the two benchmark datasets. The experimental results show that our model achieves high prediction accuracy in 10-fold cross-validation (CV) and independent set testing, outperforming existing state-of-the-art methods. Therefore, our model could serve as an effective tool for predicting AFPs.
VotePLMs-AFP:利用变压器嵌入特征和集合学习识别抗冻蛋白
抗冻蛋白(AFPs)是一类独特的生物大分子,能够保护生物体内的其他蛋白质、细胞膜和细胞结构免受冷冻条件的破坏。鉴于抗冻蛋白在生物技术、农业和医学等各个领域的重要性,人们开发了多种机器学习方法来识别抗冻蛋白。然而,由于 AFP 的复杂性和多样性,现有方法的预测性能有限。因此,迫切需要开发一种高效、快速的计算方法来准确预测 AFPs。在这项研究中,我们提出了一种基于变压器嵌入特征和集合学习的新型预测方法,用于识别 AFP,称为 VotePLMs-AFP。首先,在特征提取过程中,我们从预先训练好的蛋白质语言模型(PLMs)中提取了三种类型的特征描述符。随后,我们分析了由这三种嵌入产生的六种组合,以探索最佳特征集,并将其输入基于软投票的集合学习分类器,用于识别 AFP。最后,我们在两个基准数据集上对模型进行了评估。实验结果表明,我们的模型在 10 倍交叉验证(CV)和独立集测试中都达到了很高的预测准确率,优于现有的先进方法。因此,我们的模型可以作为预测 AFP 的有效工具。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Biochimica et biophysica acta. General subjects
生物-生化与分子生物学
CiteScore
6.40
自引率
0.00%
发文量
139
审稿时长
30 days
期刊介绍:
BBA General Subjects accepts for submission either original, hypothesis-driven studies or reviews covering subjects in biochemistry and biophysics that are considered to have general interest for a wide audience. Manuscripts with interdisciplinary approaches are especially encouraged.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


