Quantitative Analysis of Protein–Protein Equilibrium Constants in Cellular Environments Using Single-Molecule Localization Microscopy
IF 9.6
1区 材料科学
Q1 CHEMISTRY, MULTIDISCIPLINARY
引用次数: 0
Abstract
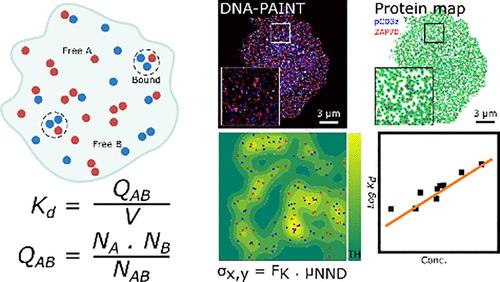
Current methods for determining equilibrium constants often operate in three-dimensional environments, which may not accurately reflect interactions with membrane-bound proteins. With our technique, based on single-molecule localization microscopy (SMLM), we directly determine protein–protein association (Ka) and dissociation (Kd) constants in cellular environments by quantifying associated and isolated molecules and their interaction area. We introduce Kernel Surface Density (ks-density,) a novel method for determining the accessible area for interacting molecules, eliminating the need for user-defined parameters. Simulation studies validate our method’s accuracy across various density and affinity conditions. Applying this technique to T cell signaling proteins, we determine the 2D association constant of T cell receptors (TCRs) in resting cells and the pseudo-3D dissociation constant of pZAP70 molecules from phosphorylated intracellular tyrosine-based activation motifs on the TCR-CD3 complex. We address challenges of multiple detection and molecular labeling efficiency. This method enhances our understanding of protein interactions in cellular environments, advancing our knowledge of complex biological processes.
利用单分子定位显微镜定量分析细胞环境中的蛋白质-蛋白质平衡常数
目前确定平衡常数的方法通常在三维环境中运行,可能无法准确反映与膜结合蛋白的相互作用。我们的技术以单分子定位显微镜(SMLM)为基础,通过量化关联分子和分离分子及其相互作用面积,直接测定细胞环境中的蛋白质-蛋白质关联常数(Ka)和解离常数(Kd)。我们引入了核表面密度(ks-density),这是一种确定相互作用分子可接触面积的新方法,无需用户定义参数。模拟研究验证了我们的方法在各种密度和亲和力条件下的准确性。将该技术应用于 T 细胞信号蛋白,我们确定了静息细胞中 T 细胞受体 (TCR) 的二维结合常数,以及 pZAP70 分子与 TCR-CD3 复合物上基于磷酸化细胞内酪氨酸的激活基团的伪三维解离常数。我们解决了多重检测和分子标记效率的难题。这种方法增强了我们对细胞环境中蛋白质相互作用的理解,促进了我们对复杂生物过程的认识。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nano Letters
工程技术-材料科学:综合
CiteScore
16.80
自引率
2.80%
发文量
1182
审稿时长
1.4 months
期刊介绍:
Nano Letters serves as a dynamic platform for promptly disseminating original results in fundamental, applied, and emerging research across all facets of nanoscience and nanotechnology. A pivotal criterion for inclusion within Nano Letters is the convergence of at least two different areas or disciplines, ensuring a rich interdisciplinary scope. The journal is dedicated to fostering exploration in diverse areas, including:
- Experimental and theoretical findings on physical, chemical, and biological phenomena at the nanoscale
- Synthesis, characterization, and processing of organic, inorganic, polymer, and hybrid nanomaterials through physical, chemical, and biological methodologies
- Modeling and simulation of synthetic, assembly, and interaction processes
- Realization of integrated nanostructures and nano-engineered devices exhibiting advanced performance
- Applications of nanoscale materials in living and environmental systems
Nano Letters is committed to advancing and showcasing groundbreaking research that intersects various domains, fostering innovation and collaboration in the ever-evolving field of nanoscience and nanotechnology.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


