Membrane structure-responsive lipid scrambling by TMEM63B to control plasma membrane lipid distribution
IF 12.5
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
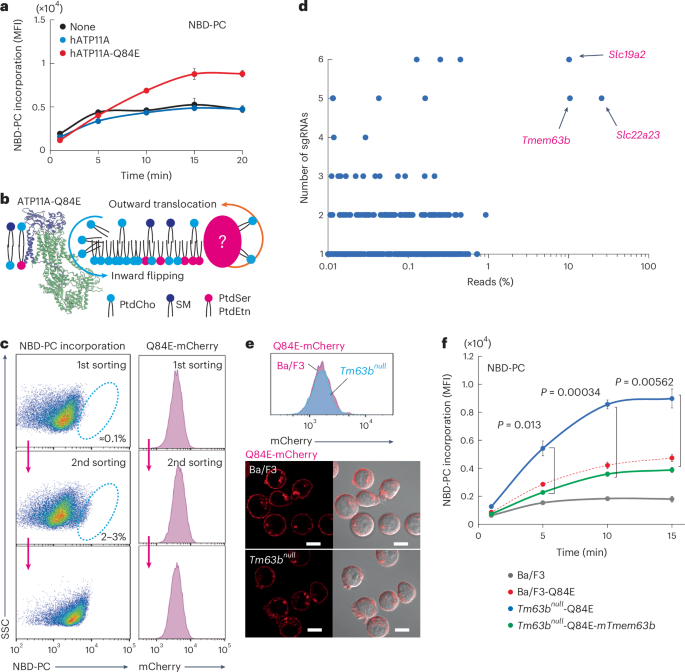
Phospholipids are asymmetrically distributed in the plasma membrane (PM), with phosphatidylcholine and sphingomyelin abundant in the outer leaflet. However, the mechanisms by which their distribution is regulated remain unclear. Here, we show that transmembrane protein 63B (TMEM63B) functions as a membrane structure-responsive lipid scramblase localized at the PM and lysosomes, activating bidirectional lipid translocation upon changes in membrane curvature and thickness. TMEM63B contains two intracellular loops with palmitoylated cysteine residue clusters essential for its scrambling function. TMEM63B deficiency alters phosphatidylcholine and sphingomyelin distributions in the PM. Persons with heterozygous mutations in TMEM63B are known to develop neurodevelopmental disorders. We show that V44M, the most frequent substitution, confers constitutive scramblase activity on TMEM63B, disrupting PM phospholipid asymmetry. We determined the cryo-electron microscopy structures of TMEM63B in its open and closed conformations, uncovering a lipid translocation pathway formed in response to changes in the membrane environment. Together, our results identify TMEM63B as a membrane structure-responsive scramblase that controls PM lipid distribution and we reveal the molecular basis for lipid scrambling and its biological importance. By combining genome-wide clustered regularly interspaced short palindromic repeats with Cas9 screening and cryo-electron microscopy structure analysis, the authors identified transmembrane protein 63B as a lipid scramblase that detects structural changes in the lipid bilayer and scrambles lipids to regulate membrane lipid distributions.

TMEM63B 的膜结构响应性脂质扰动控制质膜脂质分布
磷脂在质膜(PM)中呈不对称分布,磷脂酰胆碱和鞘磷脂在外侧小叶中含量丰富。然而,它们的分布调节机制仍不清楚。在这里,我们发现跨膜蛋白 63B(TMEM63B)具有膜结构响应性脂质扰乱酶的功能,定位于细胞质膜和溶酶体,在膜曲率和厚度发生变化时激活双向脂质转运。TMEM63B 包含两个细胞内环,其中的棕榈酰化半胱氨酸残基簇对其扰乱功能至关重要。缺乏 TMEM63B 会改变磷脂酰胆碱和鞘磷脂在 PM 中的分布。众所周知,TMEM63B 杂合子突变患者会出现神经发育障碍。我们的研究表明,V44M--最常见的取代--赋予了 TMEM63B 构成性扰乱酶活性,破坏了 PM 磷脂的不对称性。我们测定了 TMEM63B 开放构象和封闭构象的冷冻电镜结构,发现了一条响应膜环境变化而形成的脂质转运途径。总之,我们的研究结果确定了 TMEM63B 是一种控制磷脂分布的膜结构响应扰乱酶,并揭示了磷脂扰乱的分子基础及其生物学重要性。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Structural & Molecular Biology
BIOCHEMISTRY & MOLECULAR BIOLOGY-BIOPHYSICS
CiteScore
22.00
自引率
1.80%
发文量
160
审稿时长
3-8 weeks
期刊介绍:
Nature Structural & Molecular Biology is a comprehensive platform that combines structural and molecular research. Our journal focuses on exploring the functional and mechanistic aspects of biological processes, emphasizing how molecular components collaborate to achieve a particular function. While structural data can shed light on these insights, our publication does not require them as a prerequisite.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


