Bioinformatics analysis of G protein subunit gamma transduction protein 2-autophagy axis in CD11b+ dendritic cells as a potential regulator to skew airway neutrophilic inflammation in asthma endotypes
Abstract
Background
Asthma is a heterogeneous inflammatory disease with two main clinical endotypes: type 2 (T2) high and low asthma. The plasticity and autophagy in dendritic cells (DCs) influence T helper (Th)2 or Th17 differentiation to regulate asthma endotypes. Enhanced autophagy in DCs fosters Th2 differentiation in allergic environments, while reduced autophagy favors Th17 cell differentiation in sensitized and infected environments. Autophagy regulation in DCs involves interaction with various pathways like G protein-coupled receptor (GPCR), mammalian target of rapamycin (mTOR), or phosphoinositide 3-kinase (PI3K) pathway. However, specific molecules within DCs influencing asthma endotypes remain unclear.
Methods
Gene expression data series (GSE) 64896, 6858, 2276, and 55247 were obtained from gene expression omnibus (GEO) database. Differentially expressed genes (DEGs) between CD103+ and CD11b+ DCs after induction by ovalbumin (OVA) and lipopolysaccharide (LPS) were analyzed using GEO2R. DEGs were examined through Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and protein–protein interaction (PPI) analyses. The hub gene network was construct with STRING database and Cytoscape. Autophagy differences in DCs and the selected hub gene in GSE6858, GSE2276, and GSE55247 were evaluated using student t tests.
Results
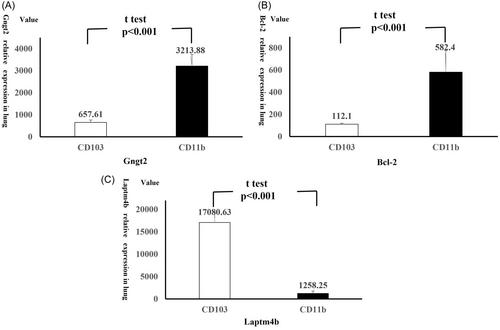
Our analysis identified 635 upregulated and 360 downregulated genes in CD11b+ DCs, compared to CD103+ DCs. These DEGs were associated with “PI3K-AKT signaling pathway,” “Ras signaling pathway,” and so forth. Thirty-five hub genes were identified, in which G protein subunit gamma transduction protein 2 (Gngt2) in CD11b+ DCs exhibited a relatively specific increase in expression associated with autophagy defects under the induction environment similar to T2 low asthma model. No significant difference was found in lung Gngt2 expression between T2 high asthma model and control group.
Conclusion
Our analysis suggested Gngt2 acted as an adapter molecule that inhibited autophagy, promoting Th17-mediated airway inflammation via the GPCR pathway in a T2 low asthma mice model. Targeting this pathway provides new asthma treatment strategies in preclinical research.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


