Multi-kernel clustering with tensor fusion on Grassmann manifold for high-dimensional genomic data
IF 4.2
3区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
The high dimensionality and noise challenges in genomic data make it difficult for traditional clustering methods. Existing multi-kernel clustering methods aim to improve the quality of the affinity matrix by learning a set of base kernels, thereby enhancing clustering performance. However, directly learning from the original base kernels presents challenges in handling errors and redundancies when dealing with high-dimensional data, and there is still a lack of feasible multi-kernel fusion strategies. To address these issues, we propose a Multi-Kernel Clustering method with Tensor fusion on Grassmann manifolds, called MKCTM. Specifically, we maximize the clustering consensus among base kernels by imposing tensor low-rank constraints to eliminate noise and redundancy. Unlike traditional kernel fusion approaches, our method fuses learned base kernels on the Grassmann manifold, resulting in a final consensus matrix for clustering. We integrate tensor learning and fusion processes into a unified optimization model and propose an effective iterative optimization algorithm for solving it. Experimental results on ten datasets, comparing against 12 popular baseline clustering methods, confirm the superiority of our approach. Our code is available at https://github.com/foureverfei/MKCTM.git.
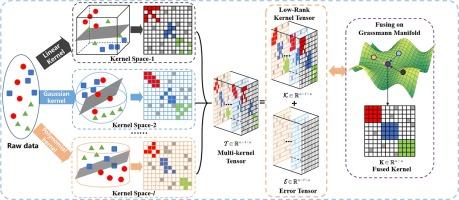
格拉斯曼流形上的多核聚类与张量融合,用于高维基因组数据
基因组数据的高维度和噪声挑战给传统聚类方法带来了困难。现有的多核聚类方法旨在通过学习一组基核来改善亲和矩阵的质量,从而提高聚类性能。然而,在处理高维数据时,直接从原始基核学习会在处理错误和冗余方面带来挑战,而且仍然缺乏可行的多核融合策略。为了解决这些问题,我们提出了一种在格拉斯曼流形上进行张量融合的多核聚类法,称为 MKCTM。具体来说,我们通过施加张量低阶约束来消除噪声和冗余,从而最大化基础内核之间的聚类共识。与传统的内核融合方法不同,我们的方法是在格拉斯曼流形上融合学习到的基础内核,从而形成最终的聚类共识矩阵。我们将张量学习和融合过程整合到一个统一的优化模型中,并提出了一种有效的迭代优化算法。在十个数据集上的实验结果与 12 种流行的基线聚类方法相比,证实了我们的方法的优越性。我们的代码见 https://github.com/foureverfei/MKCTM.git。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Methods
生物-生化研究方法
CiteScore
9.80
自引率
2.10%
发文量
222
审稿时长
11.3 weeks
期刊介绍:
Methods focuses on rapidly developing techniques in the experimental biological and medical sciences.
Each topical issue, organized by a guest editor who is an expert in the area covered, consists solely of invited quality articles by specialist authors, many of them reviews. Issues are devoted to specific technical approaches with emphasis on clear detailed descriptions of protocols that allow them to be reproduced easily. The background information provided enables researchers to understand the principles underlying the methods; other helpful sections include comparisons of alternative methods giving the advantages and disadvantages of particular methods, guidance on avoiding potential pitfalls, and suggestions for troubleshooting.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


