Structural insights into the DNA-binding mechanism of BCL11A: The integral role of ZnF6
IF 4.4
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
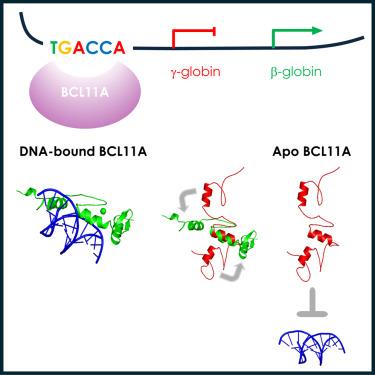
The transcription factor BCL11A is a critical regulator of the switch from fetal hemoglobin (HbF: α2γ2) to adult hemoglobin (HbA: α2β2) during development. BCL11A binds at a cognate recognition site (TGACCA) in the γ-globin gene promoter and represses its expression. DNA-binding is mediated by a triple zinc finger domain, designated ZnF456. Here, we report comprehensive investigation of ZnF456, leveraging X-ray crystallography and NMR to determine the structures in both the presence and absence of DNA. We delve into the dynamics and mode of interaction with DNA. Moreover, we discovered that the last zinc finger of BCL11A (ZnF6) plays a different role compared to ZnF4 and 5, providing a positive entropic contribution to DNA binding and γ-globin gene repression. Comprehending the DNA binding mechanism of BCL11A opens avenues for the strategic, structure-based design of novel therapeutics targeting sickle cell disease and β-thalassemia.
对 BCL11A DNA 结合机制的结构性认识:ZnF6 的重要作用
转录因子 BCL11A 是发育过程中胎儿血红蛋白(HbF:α2γ2)向成人血红蛋白(HbA:α2β2)转换的关键调节因子。BCL11A 与γ-球蛋白基因启动子中的一个同源识别位点(TGACCA)结合,并抑制其表达。DNA 结合由一个三重锌指结构域(命名为 ZnF456)介导。在此,我们报告了对 ZnF456 的全面研究,利用 X 射线晶体学和核磁共振确定了其在 DNA 存在和不存在的情况下的结构。我们深入研究了 ZnF456 与 DNA 的相互作用动力学和模式。此外,我们发现 BCL11A 的最后一个锌指(ZnF6)与 ZnF4 和 5 的作用不同,它对 DNA 结合和γ-球蛋白基因抑制有正熵贡献。了解了 BCL11A 的 DNA 结合机制,就为针对镰状细胞病和β-地中海贫血症设计基于结构的新型疗法开辟了道路。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


