Integrated strategy for high-confident global profiling of the histidine phosphoproteome
IF 5.7
2区 化学
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
Abstract
Background
Histidine phosphorylation (pHis) plays a key role in signal transduction in prokaryotes and regulates tumour initiation and progression in mammals. However, the pHis substrates and their functions are rarely known due to the lack of effective analytical strategies.
Results
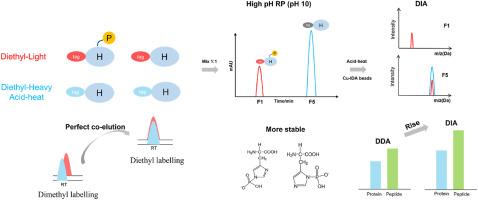
Herein, we provide a strategy for unbiased enrichment and assignment of the pHis peptides. First, the entire procedure was designed under alkaline conditions to maintain the stability of the N–P bond of pHis and high-pH reverse-phase chromatography was used to efficiently separate the pHis peptides. Second, exploiting the coelution benefits of diethyl labelling, the ratios of light- and heavy-labelled peptides were accurately quantified, and the sites of phosphorylated histidine were assigned. Finally, Cu-IDA bead enrichment and data-independent acquisition mass spectrometry analysis were used to improve the coverage of the histidine phosphoproteome. With this novel strategy, 768 and 1125 potential pHis peptides were identified from lysates of E. coli and HeLa cells, respectively. And these values represent the highest coverage of the histidine phosphoproteome for both cell types.
Significance
These data strongly support the presumption that pHis modifications are widely present in bacteria. The study provides an efficient strategy and can lead to a better understanding of pHis-modified substrates and their biological functions.

组氨酸磷酸蛋白组高可信度全球剖析综合战略
背景组氨酸磷酸化(pHis)在原核生物的信号转导中起着关键作用,并调节着哺乳动物肿瘤的发生和发展。然而,由于缺乏有效的分析策略,人们对 pHis 底物及其功能知之甚少。首先,整个过程在碱性条件下设计,以保持 pHis N-P 键的稳定性,并采用高pH反相色谱法有效分离 pHis 肽。其次,利用二乙基标记的共洗脱优势,准确量化了轻重标记肽段的比例,并确定了组氨酸磷酸化的位点。最后,利用 Cu-IDA 珠富集和数据独立采集质谱分析提高了组氨酸磷酸化蛋白质组的覆盖率。通过这种新策略,分别从大肠杆菌和 HeLa 细胞裂解液中鉴定出了 768 和 1125 个潜在的 pHis 肽。这些数据有力地支持了pHis修饰广泛存在于细菌中的推测。该研究提供了一种有效的策略,可帮助人们更好地了解 pHis 修饰的底物及其生物学功能。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Analytica Chimica Acta
化学-分析化学
CiteScore
10.40
自引率
6.50%
发文量
1081
审稿时长
38 days
期刊介绍:
Analytica Chimica Acta has an open access mirror journal Analytica Chimica Acta: X, sharing the same aims and scope, editorial team, submission system and rigorous peer review.
Analytica Chimica Acta provides a forum for the rapid publication of original research, and critical, comprehensive reviews dealing with all aspects of fundamental and applied modern analytical chemistry. The journal welcomes the submission of research papers which report studies concerning the development of new and significant analytical methodologies. In determining the suitability of submitted articles for publication, particular scrutiny will be placed on the degree of novelty and impact of the research and the extent to which it adds to the existing body of knowledge in analytical chemistry.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


