Predicting future hospital antimicrobial resistance prevalence using machine learning
IF 5.4
Q1 MEDICINE, RESEARCH & EXPERIMENTAL
引用次数: 0
Abstract
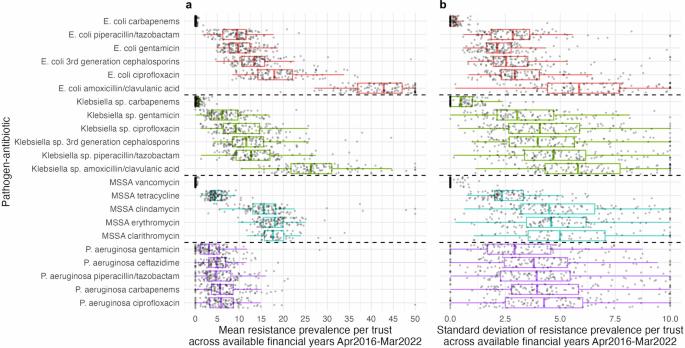
Predicting antimicrobial resistance (AMR), a top global health threat, nationwide at an aggregate hospital level could help target interventions. Using machine learning, we exploit historical AMR and antimicrobial usage to predict future AMR. Antimicrobial use and AMR prevalence in bloodstream infections in hospitals in England were obtained per hospital group (Trust) and financial year (FY, April–March) for 22 pathogen–antibiotic combinations (FY2016-2017 to FY2021-2022). Extreme Gradient Boosting (XGBoost) model predictions were compared to the previous value taken forwards, the difference between the previous two years taken forwards and linear trend forecasting (LTF). XGBoost feature importances were calculated to aid interpretability. Here we show that XGBoost models achieve the best predictive performance. Relatively limited year-to-year variability in AMR prevalence within Trust–pathogen–antibiotic combinations means previous value taken forwards also achieves a low mean absolute error (MAE), similar to or slightly higher than XGBoost. Using the difference between the previous two years taken forward or LTF performs consistently worse. XGBoost considerably outperforms all other methods in Trusts with a larger change in AMR prevalence from FY2020-2021 (last training year) to FY2021-2022 (held-out test set). Feature importance values indicate that besides historical resistance to the same pathogen–antibiotic combination as the outcome, complex relationships between resistance in different pathogens to the same antibiotic/antibiotic class and usage are exploited for predictions. These are generally among the top ten features ranked according to their mean absolute SHAP values. Year-to-year resistance has generally changed little within Trust–pathogen–antibiotic combinations. In those with larger changes, XGBoost models can improve predictions, enabling informed decisions, efficient resource allocation, and targeted interventions. Antibiotics play an important role in treating serious bacterial infections. However, with the increased usage of antibiotics, they are becoming less effective. In our study, we use machine learning to learn from past antibiotic resistance and usage in order to predict what resistance will look like in the future. Different hospitals across England have very different resistance levels, however, within each hospital, these levels remain stable over time. When larger changes in resistance occurred over time in individual hospitals, our methods were able to predict these. Understanding how much resistance there is in hospital populations, and what may occur in the future can help determine where resources and interventions should be directed. Vihta et al. use past hospital data including bloodstream infection cases, susceptibilities, and antimicrobial use to predict future resistance prevalence. Machine learning can improve the accuracy of predictions potentially impacting interventions.
利用机器学习预测未来医院的抗菌药耐药性流行率。
背景:抗菌药物耐药性(AMR)是全球最大的健康威胁,在全国范围内对医院进行综合预测有助于确定干预措施的目标。通过机器学习,我们利用历史上的抗菌药物耐药性和抗菌药物使用情况来预测未来的抗菌药物耐药性:方法:我们按医院集团(信托)和财政年度(FY,4 月至 3 月)获得了 22 种病原体-抗生素组合(2016-2017 财年至 2021-2022 财年)的抗菌药物使用情况和英格兰医院血流感染中的 AMR 流行率。极端梯度提升(XGBoost)模型的预测结果与前值、前两年的差值和线性趋势预测(LTF)进行了比较。为了帮助解释,还计算了 XGBoost 的特征导入值:结果:我们在此表明,XGBoost 模型实现了最佳预测性能。在信托基金-病原体-抗生素组合中,AMR 流行率的年际变化相对有限,这意味着前值也能达到较低的平均绝对误差 (MAE),与 XGBoost 相似或略高于 XGBoost。使用前两年前移值或 LTF 之间的差值则一直表现较差。在从 2020-2021 财年(最后一个训练年)到 2021-2022 财年(保持不变的测试集)AMR 流行率变化较大的信托机构中,XGBoost 明显优于所有其他方法。特征重要性值表明,除了将历史上对同一病原体-抗生素组合的耐药性作为结果外,还利用了不同病原体对同一抗生素/抗生素类别的耐药性和使用情况之间的复杂关系来进行预测。根据其平均绝对 SHAP 值,这些特征通常排在前十位:结论:在信托基金-病原体-抗生素组合中,逐年的耐药性变化一般不大。对于那些变化较大的组合,XGBoost 模型可以改进预测,从而做出明智的决策、高效的资源分配和有针对性的干预。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


