Comprehensive analysis of prognostic and immunological role of basement membrane-related genes in soft tissue sarcoma
Abstract
Background
Soft tissue sarcoma (STS) represents highly multifarious malignant tumors that often occur in adolescents and have a poor prognosis. The basement membrane, as an ancient cellular matrix, was recently proven to play a vital role in developing abundant tumors. The relationship between basement membrane-related genes and STS remains unknown.
Methods
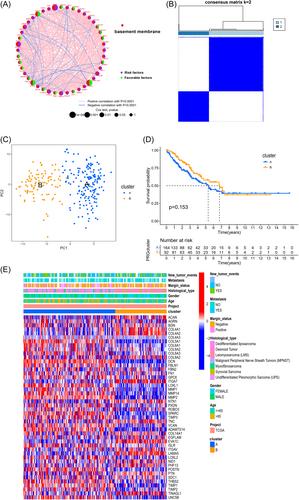
Consensus clustering was employed to identify subgroups related to differentially expressed basement membrane-related genes. Cox and least absolute shrinkage and selection operator regression analyses were utilized to construct this novel signature. Then, we established a nomogram and calibration curve, including the risk score and available clinical characteristics. Finally, we carried out functional enrichment analysis and immune microenvironment analysis to investigate enriched pathways and the tumor immune microenvironment related to the novel signature.
Results
A prognostic predictive signature consisting of eight basement membrane-related genes was established. Kaplan–Meier survival curves demonstrated that the patients in the high-risk group had a poor prognosis. Independent analysis illustrated that this risk model could be an independent prognostic predictor. We validated the accuracy of our signature in the validation data set. In addition, gene set enrichment analysis and immune microenvironment analysis showed that patients with low-risk scores were enriched in some pathways associated with immunity. Finally, in vitro experiments showed significantly differential expression levels of these signature genes in STS cells and PSAT1 could promote the malignant behavior of STS.
Conclusions
The novel signature is a promising prognostic predictor for STS. The present study may improve the prognosis and enhance individualized treatment for STS in the future.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


