scNanoSeq-CUT&Tag: a single-cell long-read CUT&Tag sequencing method for efficient chromatin modification profiling within individual cells
IF 32.1
1区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
Chromatin modifications are fundamental epigenetic marks that determine genome functions, but it remains challenging to profile those of repetitive elements and complex genomic regions. Here, we develop scNanoSeq-CUT&Tag, a streamlined method, by adapting modified cleavage under targets and tagmentation (CUT&Tag) to the nanopore sequencing platform for genome-wide chromatin modification profiling within individual cells. We show that scNanoSeq-CUT&Tag can accurately profile histone marks and transcription factor occupancy patterns at single-cell resolution as well as distinguish different cell types. scNanoSeq-CUT&Tag efficiently maps the allele-specific chromatin modifications and allows analysis of their neighboring region co-occupancy patterns within individual cells. Moreover, scNanoSeq-CUT&Tag can accurately detect chromatin modifications for individual copies of repetitive elements in both human and mouse genomes. Overall, we prove that scNanoSeq-CUT&Tag is a valuable single-cell tool for efficiently profiling histone marks and transcription factor occupancies, especially for previously poorly studied complex genomic regions and blacklist genomic regions. This work presents scNanoSeq-CUT&Tag, using long-read sequencing to profile histone modifications and DNA-binding proteins at the single-cell level.
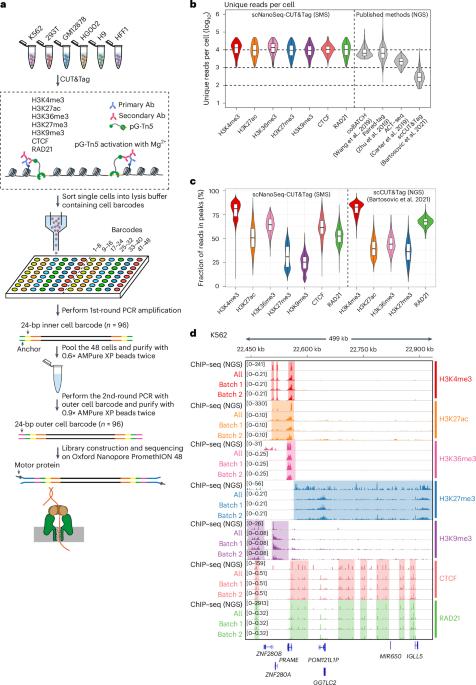
scNanoSeq-CUT&Tag:一种单细胞长线程 CUT&Tag 测序方法,用于高效分析单个细胞内的染色质修饰。
染色质修饰是决定基因组功能的基本表观遗传标记,但对重复元件和复杂基因组区域的染色质修饰进行剖析仍是一项挑战。在这里,我们开发了 scNanoSeq-CUT&Tag,这是一种简化的方法,它将目标下的改良裂解和标记(CUT&Tag)应用于纳米孔测序平台,用于单个细胞内的全基因组染色质修饰谱分析。我们的研究表明,scNanoSeq-CUT&Tag 能以单细胞分辨率准确描绘组蛋白标记和转录因子占据模式,并能区分不同的细胞类型。scNanoSeq-CUT&Tag 能有效描绘等位基因特异性染色质修饰,并能分析它们在单个细胞内的邻近区域共占据模式。此外,scNanoSeq-CUT&Tag 还能准确检测人类和小鼠基因组中重复元件单个拷贝的染色质修饰。总之,我们证明了 scNanoSeq-CUT&Tag 是高效剖析组蛋白标记和转录因子占位的重要单细胞工具,尤其适用于以前研究较少的复杂基因组区域和黑名单基因组区域。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Methods
生物-生化研究方法
CiteScore
58.70
自引率
1.70%
发文量
326
审稿时长
1 months
期刊介绍:
Nature Methods is a monthly journal that focuses on publishing innovative methods and substantial enhancements to fundamental life sciences research techniques. Geared towards a diverse, interdisciplinary readership of researchers in academia and industry engaged in laboratory work, the journal offers new tools for research and emphasizes the immediate practical significance of the featured work. It publishes primary research papers and reviews recent technical and methodological advancements, with a particular interest in primary methods papers relevant to the biological and biomedical sciences. This includes methods rooted in chemistry with practical applications for studying biological problems.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


