Rare variant analyses in 51,256 type 2 diabetes cases and 370,487 controls reveal the pathogenicity spectrum of monogenic diabetes genes
IF 31.7
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
Abstract
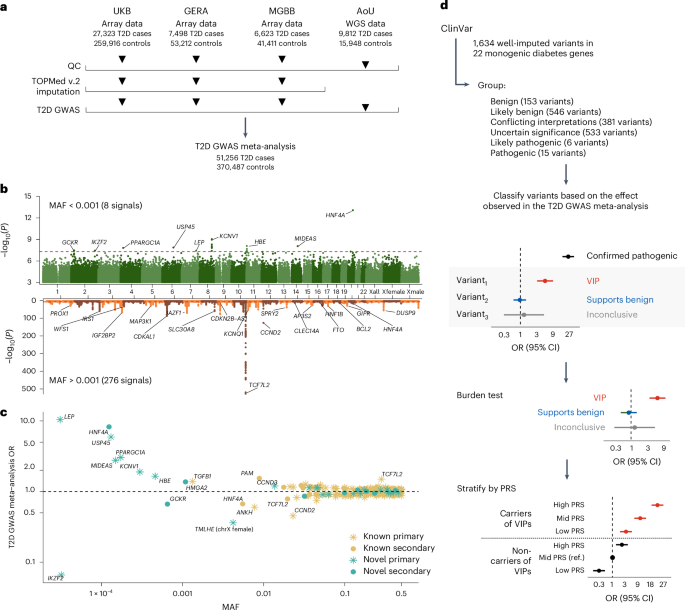
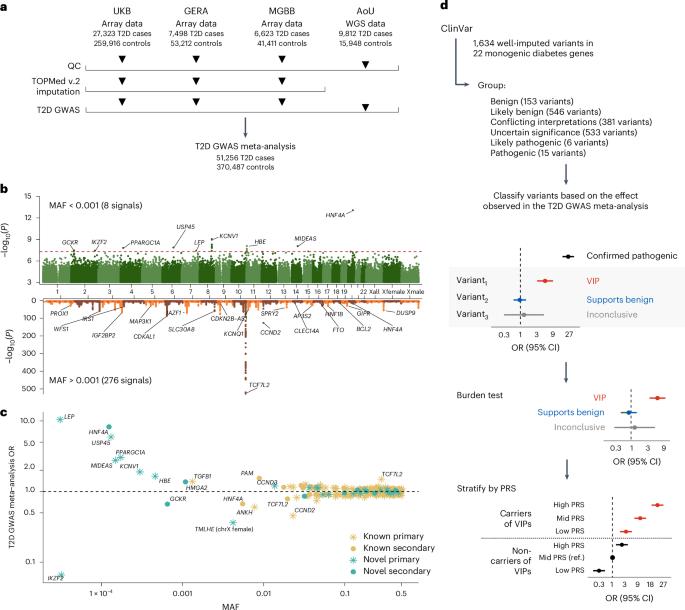
Type 2 diabetes (T2D) genome-wide association studies (GWASs) often overlook rare variants as a result of previous imputation panels’ limitations and scarce whole-genome sequencing (WGS) data. We used TOPMed imputation and WGS to conduct the largest T2D GWAS meta-analysis involving 51,256 cases of T2D and 370,487 controls, targeting variants with a minor allele frequency as low as 5 × 10−5. We identified 12 new variants, including a rare African/African American-enriched enhancer variant near the LEP gene (rs147287548), associated with fourfold increased T2D risk. We also identified a rare missense variant in HNF4A (p.Arg114Trp), associated with eightfold increased T2D risk, previously reported in maturity-onset diabetes of the young with reduced penetrance, but observed here in a T2D GWAS. We further leveraged these data to analyze 1,634 ClinVar variants in 22 genes related to monogenic diabetes, identifying two additional rare variants in HNF1A and GCK associated with fivefold and eightfold increased T2D risk, respectively, the effects of which were modified by the individual’s polygenic risk score. For 21% of the variants with conflicting interpretations or uncertain significance in ClinVar, we provided support of being benign based on their lack of association with T2D. Our work provides a framework for using rare variant GWASs to identify large-effect variants and assess variant pathogenicity in monogenic diabetes genes. Rare variant analyses identify a new type 2 diabetes risk allele near the LEP gene, which encodes leptin, and other risk alleles of intermediate penetrance in genes previously implicated in monogenic forms of diabetes.
对 51,256 例 2 型糖尿病病例和 370,487 例对照病例进行的罕见变异分析揭示了单基因糖尿病基因的致病范围
由于以往估算板的局限性和全基因组测序(WGS)数据的稀缺,2 型糖尿病(T2D)全基因组关联研究(GWAS)经常会忽略罕见变异。我们利用 TOPMed 估算和 WGS 进行了最大规模的 T2D GWAS 元分析,涉及 51,256 例 T2D 病例和 370,487 例对照,目标是小等位基因频率低至 5 × 10-5 的变异。我们发现了 12 个新变异,包括 LEP 基因附近一个罕见的非洲/非裔美国人丰富的增强子变异(rs147287548),它与 T2D 风险增加四倍有关。我们还在 HNF4A(p.Arg114Trp)中发现了一个罕见的错义变异,它与 T2D 风险增加 8 倍有关。我们进一步利用这些数据分析了与单基因糖尿病相关的 22 个基因中的 1,634 个 ClinVar 变异,在 HNF1A 和 GCK 中又发现了两个分别与 T2D 风险增加五倍和八倍相关的罕见变异,它们的影响会因个体的多基因风险评分而改变。在 ClinVar 中,有 21% 的变异具有相互矛盾的解释或不确定的意义,我们根据它们与 T2D 缺乏关联的情况,证明它们是良性的。我们的工作为利用罕见变异GWAS鉴定大效应变异和评估单基因糖尿病基因的变异致病性提供了一个框架。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature genetics
生物-遗传学
CiteScore
43.00
自引率
2.60%
发文量
241
审稿时长
3 months
期刊介绍:
Nature Genetics publishes the very highest quality research in genetics. It encompasses genetic and functional genomic studies on human and plant traits and on other model organisms. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation.
Integrative genetic topics comprise, but are not limited to:
-Genes in the pathology of human disease
-Molecular analysis of simple and complex genetic traits
-Cancer genetics
-Agricultural genomics
-Developmental genetics
-Regulatory variation in gene expression
-Strategies and technologies for extracting function from genomic data
-Pharmacological genomics
-Genome evolution
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


