Structural mechanism of HP1⍺-dependent transcriptional repression and chromatin compaction
IF 4.4
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
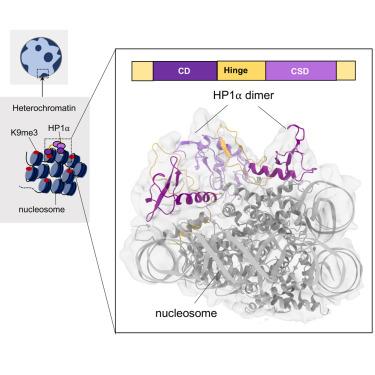
Heterochromatin protein 1 (HP1) plays a central role in establishing and maintaining constitutive heterochromatin. However, the mechanisms underlying HP1-nucleosome interactions and their contributions to heterochromatin functions remain elusive. Here, we present the cryoelectron microscopy (cryo-EM) structure of an HP1α dimer bound to an H2A.Z-nucleosome, revealing two distinct HP1α-nucleosome interfaces. The primary HP1α binding site is located at the N terminus of histone H3, specifically at the trimethylated lysine 9 (K9me3) region, while a secondary binding site is situated near histone H2B, close to nucleosome superhelical location 4 (SHL4). Our biochemical data further demonstrates that HP1α binding influences the dynamics of DNA on the nucleosome. It promotes DNA unwrapping near the nucleosome entry and exit sites while concurrently restricting DNA accessibility in the vicinity of SHL4. Our study offers a model for HP1α-mediated heterochromatin maintenance and gene silencing. It also sheds light on the H3K9me-independent role of HP1 in responding to DNA damage.
HP1⍺ 依赖性转录抑制和染色质压实的结构机制
异染色质蛋白 1(HP1)在建立和维持组成型异染色质中发挥着核心作用。然而,HP1-核小体相互作用的机制及其对异染色质功能的贡献仍然难以捉摸。在这里,我们展示了与 H2A.Z 核小体结合的 HP1α 二聚体的冷冻电子显微镜(cryo-EM)结构,揭示了两个不同的 HP1α 核小体界面。主要的HP1α结合位点位于组蛋白H3的N末端,特别是三甲基化赖氨酸9(K9me3)区域,而次要结合位点位于组蛋白H2B附近,靠近核小体超螺旋位置4(SHL4)。我们的生化数据进一步证明,HP1α的结合会影响核小体上DNA的动态。它促进了核小体入口和出口位点附近的DNA解链,同时限制了SHL4附近DNA的可及性。我们的研究为HP1α介导的异染色质维持和基因沉默提供了一个模型。这项研究还揭示了HP1在应对DNA损伤时与H3K9me无关的作用。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


