SNRPB2 in the pan-cancer landscape: A bioinformatics exploration and validation in hepatocellular carcinoma
IF 4.4
2区 生物学
Q2 CELL BIOLOGY
引用次数: 0
Abstract
Aberrant splicing is a significant contributor to gene expression abnormalities in cancer. SNRPB2, a component of U2 small nuclear ribonucleoprotein particles (snRNPs), contributes to the assembly of the spliceosome, the molecular machinery responsible for splicing. To date, few studies have investigated the role of SNRPB2 in tumorigenesis. We examined data sourced from various public databases, such as The Cancer Genome Atlas(TCGA), the Clinical Proteomic Tumor Analysis Consortium(CPTAC), and Gene Expression Omnibus(GEO). Our investigation included gene expression, genomic and epigenomic scrutiny, gene set enrichment assessment(GSEA), and immune cell infiltration evaluation. Furthermore, we performed empirical validation to ascertain the impact of SNRPB2 suppression on the proliferation and migration of liver cancer cells. Analysis of gene expression revealed widespread upregulation of SNRPB2 across a spectrum of cancer types, with heightened levels of SNRPB2 expression in numerous tumors linked to unfavorable prognosis. Genomic and epigenomic assessments revealed connections between SNRPB2 expression and variations in SNRPB2 copy number, DNA methylation patterns, and RNA modifications. Through gene set enrichment analysis, the involvement of SNRPB2 in vital biological processes and pathways related to cancer was identified. Furthermore, scrutiny of immune cell infiltration suggested a potential relationship between SNRPB2 and the tumor microenvironment, which was reinforced by multiple single-cell sequencing profiles. Subsequent experimental validation revealed that silencing SNRPB2 effectively impeded the proliferation and migration of liver cancer cells. Taken together, these findings underscore the prospective utility of SNRPB2 as a prognostic biomarker and a promising candidate for immunotherapy in cancer. It is necessary to engage in additional exploration into its underlying mechanisms and clinical treatment potential.
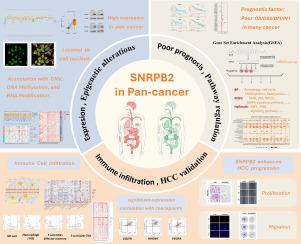
泛癌症中的 SNRPB2:肝细胞癌中的生物信息学探索与验证。
剪接异常是导致癌症基因表达异常的一个重要因素。SNRPB2是U2小核核糖核蛋白颗粒(snRNPs)的一个组成部分,它有助于剪接体的组装,剪接体是负责剪接的分子机制。迄今为止,很少有研究调查 SNRPB2 在肿瘤发生中的作用。我们研究了来自各种公共数据库的数据,如癌症基因组图谱(TCGA)、临床蛋白质组肿瘤分析联盟(CPTAC)和基因表达总库(GEO)。我们的研究包括基因表达、基因组和表观基因组审查、基因组富集评估(GSEA)和免疫细胞浸润评估。此外,我们还进行了经验验证,以确定抑制 SNRPB2 对肝癌细胞增殖和迁移的影响。基因表达分析表明,SNRPB2在多种癌症类型中广泛上调,许多肿瘤中SNRPB2表达水平的升高与预后不良有关。基因组和表观基因组评估显示,SNRPB2的表达与SNRPB2拷贝数、DNA甲基化模式和RNA修饰的变化有关。通过基因组富集分析,确定了 SNRPB2 参与与癌症有关的重要生物过程和途径。此外,对免疫细胞浸润的研究表明,SNRPB2 与肿瘤微环境之间存在潜在的关系,而多个单细胞测序图谱又进一步证实了这一点。随后的实验验证表明,沉默 SNRPB2 能有效抑制肝癌细胞的增殖和迁移。综上所述,这些发现强调了SNRPB2作为预后生物标志物和癌症免疫疗法候选药物的前景。我们有必要对其潜在机制和临床治疗潜力进行进一步的探索。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Cellular signalling
生物-细胞生物学
CiteScore
8.40
自引率
0.00%
发文量
250
审稿时长
27 days
期刊介绍:
Cellular Signalling publishes original research describing fundamental and clinical findings on the mechanisms, actions and structural components of cellular signalling systems in vitro and in vivo.
Cellular Signalling aims at full length research papers defining signalling systems ranging from microorganisms to cells, tissues and higher organisms.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


