Mechanism of phospho-Ubls’ specificity and conformational changes that regulate Parkin activity
IF 4.4
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
PINK1 and Parkin mutations lead to the early onset of Parkinson’s disease. PINK1-mediated phosphorylation of ubiquitin (Ub), ubiquitin-like protein (NEDD8), and ubiquitin-like (Ubl) domain of Parkin activate autoinhibited Parkin E3 ligase. The mechanism of various phospho-Ubls’ specificity and conformational changes leading to Parkin activation remain elusive. Herein, we show that compared to Ub, NEDD8 is a more robust binder and activator of Parkin. Structures and biophysical/biochemical data reveal specific recognition and underlying mechanisms of pUb/pNEDD8 and pUbl domain binding to the RING1 and RING0 domains, respectively. Also, pUb/pNEDD8 binding in the RING1 pocket promotes allosteric conformational changes in Parkin’s catalytic domain (RING2), leading to Parkin activation. Furthermore, Parkinson’s disease mutation K211N in the RING0 domain was believed to perturb Parkin activation due to loss of pUb binding. However, our data reveal allosteric conformational changes due to N211 that lock RING2 with RING0 to inhibit Parkin activity without disrupting pNEDD8/pUb binding.
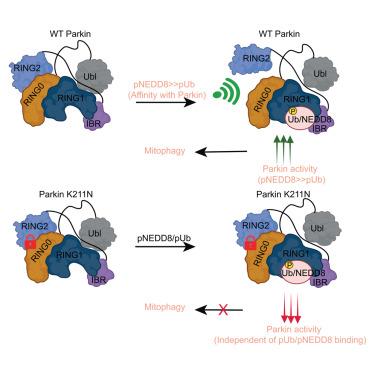
磷酸化-Ubls 的特异性和构象变化调节 Parkin 活性的机制
PINK1 和 Parkin 基因突变会导致帕金森病的早期发病。PINK1 介导的泛素(Ub)、泛素样蛋白(NEDD8)和 Parkin 的泛素样(Ubl)结构域的磷酸化激活了自身抑制的 Parkin E3 连接酶。各种磷酸化-Ubls的特异性和构象变化导致Parkin活化的机制仍未确定。在这里,我们发现与 Ub 相比,NEDD8 是一种更强大的 Parkin 结合剂和激活剂。结构和生物物理/生化数据揭示了 pUb/pNEDD8 和 pUbl 结构域分别与 RING1 和 RING0 结构域结合的特异性识别和基本机制。此外,pUb/pNEDD8 在 RING1 口袋中的结合促进了 Parkin 催化结构域(RING2)的异构构象变化,从而导致 Parkin 激活。此外,帕金森病患者认为 RING0 结构域中的 K211N 突变会因失去 pUb 结合而扰乱 Parkin 的激活。然而,我们的数据揭示了 N211 导致的异构构象变化,这种变化将 RING2 与 RING0 锁定,从而抑制了 Parkin 的活性,而不会破坏 pNEDD8/pUb 的结合。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


