Multiomic profiling identifies predictors of survival in African American patients with acute myeloid leukemia
IF 31.7
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
Abstract
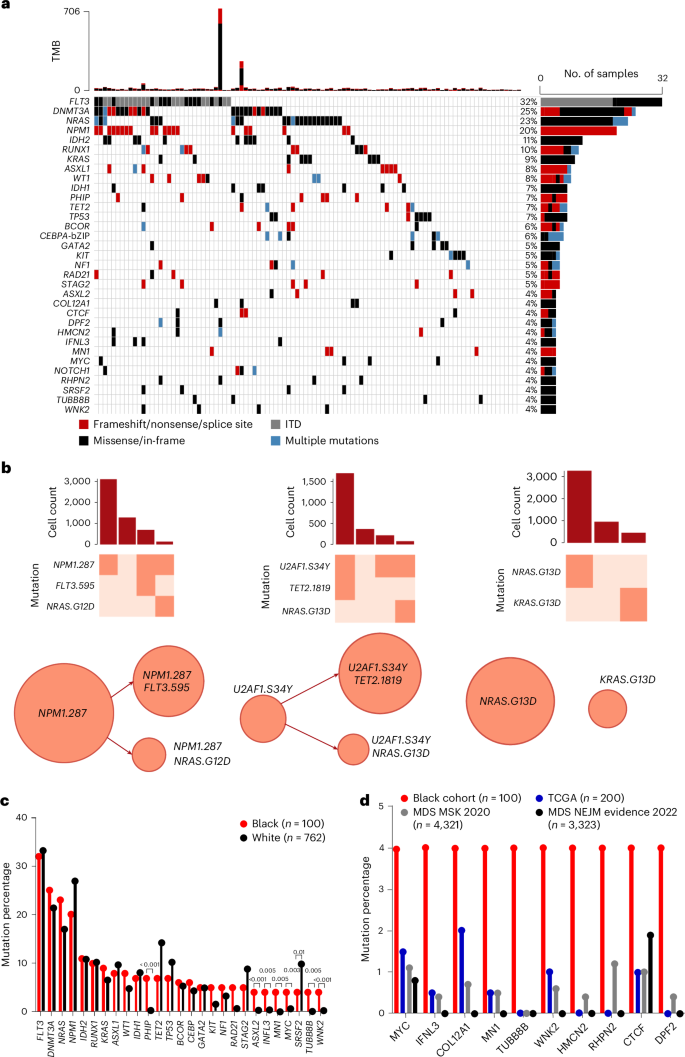
Genomic profiles and prognostic biomarkers in patients with acute myeloid leukemia (AML) from ancestry-diverse populations are underexplored. We analyzed the exomes and transcriptomes of 100 patients with AML with genomically confirmed African ancestry (Black; Alliance) and compared their somatic mutation frequencies with those of 323 self-reported white patients with AML, 55% of whom had genomically confirmed European ancestry (white; BeatAML). Here we find that 73% of 162 gene mutations recurrent in Black patients, including a hitherto unreported PHIP alteration detected in 7% of patients, were found in one white patient or not detected. Black patients with myelodysplasia-related AML were younger than white patients suggesting intrinsic and/or extrinsic dysplasia-causing stressors. On multivariable analyses of Black patients, NPM1 and NRAS mutations were associated with inferior disease-free and IDH1 and IDH2 mutations with reduced overall survival. Inflammatory profiles, cell type distributions and transcriptional profiles differed between Black and white patients with NPM1 mutations. Incorporation of ancestry-specific risk markers into the 2022 European LeukemiaNet genetic risk stratification changed risk group assignment for one-third of Black patients and improved their outcome prediction. Analysis of exomes and transcriptomes from 100 African American patients with acute myeloid leukemia identifies ancestry-related variation in mutation profiles and survival. Refined risk classification suggests clinical relevance of these ancestry-associated differences.
多基因组分析确定非裔美国人急性髓性白血病患者的生存预测指标
对来自不同血统人群的急性髓性白血病(AML)患者的基因组图谱和预后生物标志物的研究还很不够。我们分析了 100 名经基因组学证实具有非洲血统(黑人;Alliance)的急性髓性白血病患者的外显子组和转录组,并将他们的体细胞突变频率与 323 名自我报告的白人急性髓性白血病患者的体细胞突变频率进行了比较,其中 55% 的患者经基因组学证实具有欧洲血统(白人;BeatAML)。我们在此发现,在黑人患者中复发的 162 个基因突变中,73%(包括在 7% 患者中检测到的迄今未报道的 PHIP 变异)在一名白人患者中发现或未检测到。骨髓增生异常相关急性髓细胞白血病的黑人患者比白人患者年轻,这表明有内在和/或外在导致增生异常的压力因素。对黑人患者进行多变量分析后发现,NPM1和NRAS突变与无病生存率降低有关,而IDH1和IDH2突变与总生存率降低有关。NPM1突变的黑人和白人患者的炎症特征、细胞类型分布和转录特征各不相同。在2022年欧洲白血病网络遗传风险分层中纳入血统特异性风险标记,改变了三分之一黑人患者的风险组别分配,并改善了他们的预后。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature genetics
生物-遗传学
CiteScore
43.00
自引率
2.60%
发文量
241
审稿时长
3 months
期刊介绍:
Nature Genetics publishes the very highest quality research in genetics. It encompasses genetic and functional genomic studies on human and plant traits and on other model organisms. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation.
Integrative genetic topics comprise, but are not limited to:
-Genes in the pathology of human disease
-Molecular analysis of simple and complex genetic traits
-Cancer genetics
-Agricultural genomics
-Developmental genetics
-Regulatory variation in gene expression
-Strategies and technologies for extracting function from genomic data
-Pharmacological genomics
-Genome evolution
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


