Molecular basis of plastoquinone reduction in plant cytochrome b6f
IF 15.8
1区 生物学
Q1 PLANT SCIENCES
引用次数: 0
Abstract
A multi-subunit enzyme, cytochrome b6f (cytb6f), provides the crucial link between photosystems I and II in the photosynthetic membranes of higher plants, transferring electrons between plastoquinone (PQ) and plastocyanin. The atomic structure of cytb6f is known, but its detailed catalytic mechanism remains elusive. Here we present cryogenic electron microscopy structures of spinach cytb6f at 1.9 Å and 2.2 Å resolution, revealing an unexpected orientation of the substrate PQ in the haem ligand niche that forms the PQ reduction site (Qn). PQ, unlike Qn inhibitors, is not in direct contact with the haem. Instead, a water molecule is coordinated by one of the carbonyl groups of PQ and can act as the immediate proton donor for PQ. In addition, we identify water channels that connect Qn with the aqueous exterior of the enzyme, suggesting that the binding of PQ in Qn displaces water through these channels. The structures confirm large movements of the head domain of the iron–sulfur protein (ISP-HD) towards and away from the plastoquinol oxidation site (Qp) and define the unique position of ISP-HD when a Qp inhibitor (2,5-dibromo-3-methyl-6-isopropylbenzoquinone) is bound. This work identifies key conformational states of cytb6f, highlights fundamental differences between substrates and inhibitors and proposes a quinone–water exchange mechanism. Cryo-EM structures of cytb6f with quinone bound in the Qn site allow the identification of specific proton channels and reveal differences to known inhibitors. Nearby water channels imply a ‘piston-like’ mechanism during catalytic turnover.

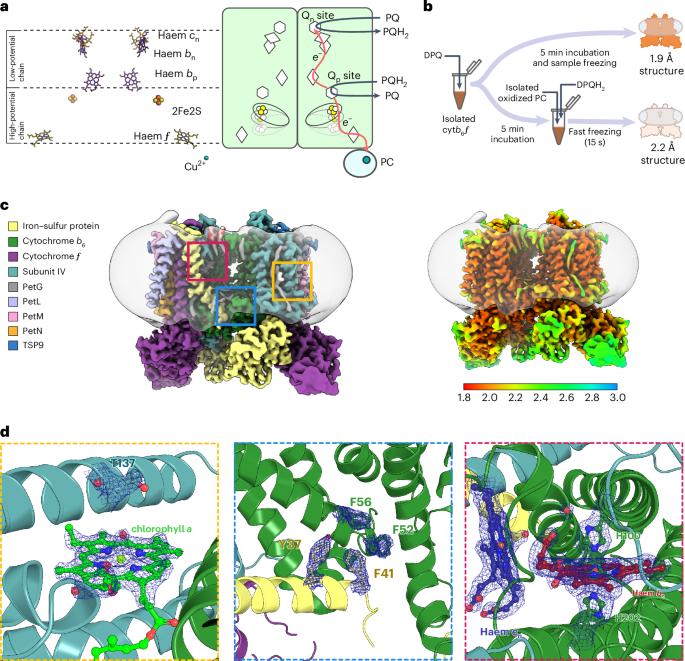
植物细胞色素 b6f 中质子醌还原的分子基础
一种多亚基酶--细胞色素 b6f(cytb6f)--是高等植物光合膜中光合系统 I 和 II 之间的关键纽带,它在质醌(PQ)和质花青素之间传递电子。细胞b6f的原子结构已知,但其详细的催化机理仍不清楚。在这里,我们以 1.9 Å 和 2.2 Å 的分辨率展示了菠菜 cytb6f 的低温电子显微镜结构,揭示了底物 PQ 在形成 PQ 还原位点(Qn)的血红素配体位点中的意外取向。与 Qn 抑制剂不同,PQ 并不与血红素直接接触。相反,一个水分子与 PQ 的一个羰基配位,可作为 PQ 的直接质子供体。此外,我们还发现了连接 Qn 与酶的水外部的水通道,这表明 Qn 中 PQ 的结合会通过这些通道置换出水。这些结构证实了铁硫蛋白头部结构域(ISP-HD)向和远离质醌氧化位点(Qp)的大量运动,并确定了当 Qp 抑制剂(2,5-二溴-3-甲基-6-异丙基苯醌)结合时 ISP-HD 的独特位置。这项工作确定了细胞b6f的关键构象状态,突出了底物和抑制剂之间的根本差异,并提出了一种醌水交换机制。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Nature Plants
PLANT SCIENCES-
CiteScore
25.30
自引率
2.20%
发文量
196
期刊介绍:
Nature Plants is an online-only, monthly journal publishing the best research on plants — from their evolution, development, metabolism and environmental interactions to their societal significance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


