Mechanisms and regulation of substrate degradation by the 26S proteasome
IF 81.3
1区 生物学
Q1 CELL BIOLOGY
引用次数: 0
Abstract
The 26S proteasome is involved in degrading and regulating the majority of proteins in eukaryotic cells, which requires a sophisticated balance of specificity and promiscuity. In this Review, we discuss the principles that underly substrate recognition and ATP-dependent degradation by the proteasome. We focus on recent insights into the mechanisms of conventional ubiquitin-dependent and ubiquitin-independent protein turnover, and discuss the plethora of modulators for proteasome function, including substrate-delivering cofactors, ubiquitin ligases and deubiquitinases that enable the targeting of a highly diverse substrate pool. Furthermore, we summarize recent progress in our understanding of substrate processing upstream of the 26S proteasome by the p97 protein unfoldase. The advances in our knowledge of proteasome structure, function and regulation also inform new strategies for specific inhibition or harnessing the degradation capabilities of the proteasome for the treatment of human diseases, for instance, by using proteolysis targeting chimera molecules or molecular glues. Most proteins are degraded and regulated by the 26S proteasome. A balance of specificity and promiscuity underlies substrate recognition and ATP-dependent degradation, which can occur in a ubiquitin-dependent or ubiquitin-independent manner. Recent insights into the mechanisms and regulation of substrate delivery, processing and degradation will inform new strategies for the treatment of human diseases.

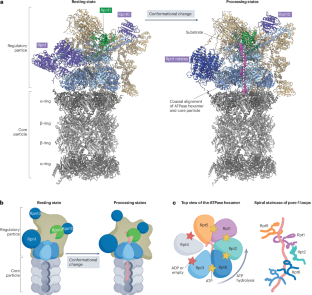
26S 蛋白酶体降解底物的机制和调控
26S 蛋白酶体参与降解和调节真核细胞中的大多数蛋白质,这需要在特异性和杂交性之间取得复杂的平衡。在本综述中,我们将讨论蛋白酶体识别底物和 ATP 依赖性降解的原理。我们重点探讨了对传统泛素依赖型和泛素非依赖型蛋白质周转机制的最新见解,并讨论了蛋白酶体功能的大量调节剂,包括底物递送辅助因子、泛素连接酶和去泛素酶,它们使蛋白酶体能够靶向高度多样化的底物库。此外,我们还总结了最近在了解 26S 蛋白酶体上游 p97 蛋白未折叠酶处理底物方面取得的进展。我们对蛋白酶体结构、功能和调控认识的进步,也为特异性抑制或利用蛋白酶体降解能力治疗人类疾病的新策略提供了依据,例如通过使用蛋白水解靶向嵌合体分子或分子胶。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
CiteScore
173.60
自引率
0.50%
发文量
118
审稿时长
6-12 weeks
期刊介绍:
Nature Reviews Molecular Cell Biology is a prestigious journal that aims to be the primary source of reviews and commentaries for the scientific communities it serves. The journal strives to publish articles that are authoritative, accessible, and enriched with easily understandable figures, tables, and other display items. The goal is to provide an unparalleled service to authors, referees, and readers, and the journal works diligently to maximize the usefulness and impact of each article. Nature Reviews Molecular Cell Biology publishes a variety of article types, including Reviews, Perspectives, Comments, and Research Highlights, all of which are relevant to molecular and cell biologists. The journal's broad scope ensures that the articles it publishes reach the widest possible audience.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


