MHC Hammer reveals genetic and non-genetic HLA disruption in cancer evolution
IF 31.7
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
Abstract
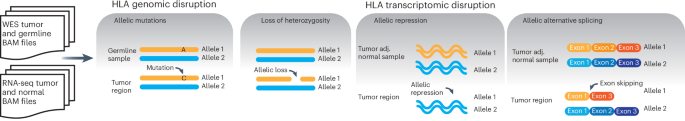
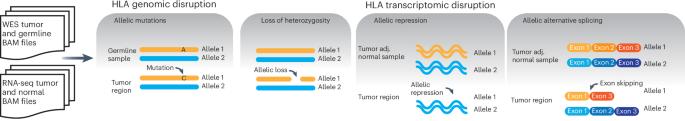
Disruption of the class I human leukocyte antigen (HLA) molecules has important implications for immune evasion and tumor evolution. We developed major histocompatibility complex loss of heterozygosity (LOH), allele-specific mutation and measurement of expression and repression (MHC Hammer). We identified extensive variability in HLA allelic expression and pervasive HLA alternative splicing in normal lung and breast tissue. In lung TRACERx and lung and breast TCGA cohorts, 61% of lung adenocarcinoma (LUAD), 76% of lung squamous cell carcinoma (LUSC) and 35% of estrogen receptor-positive (ER+) cancers harbored class I HLA transcriptional repression, while HLA tumor-enriched alternative splicing occurred in 31%, 11% and 15% of LUAD, LUSC and ER+ cancers. Consistent with the importance of HLA dysfunction in tumor evolution, in LUADs, HLA LOH was associated with metastasis and LUAD primary tumor regions seeding a metastasis had a lower effective neoantigen burden than non-seeding regions. These data highlight the extent and importance of HLA transcriptomic disruption, including repression and alternative splicing in cancer evolution. Major histocompatibility complex (MHC) loss of heterozygosity, allele-specific mutation and measurement of expression and repression (MHC Hammer) detects disruption to human leukocyte antigens due to mutations, loss of heterogeneity, altered gene expression or alternative splicing. Applied to lung and breast cancer datasets, the tool shows that these aberrations are common across cancer and can have clinical implications.
MHC Hammer 揭示了癌症进化过程中的遗传和非遗传 HLA 干扰
一类人类白细胞抗原(HLA)分子的破坏对免疫逃避和肿瘤进化有重要影响。我们开发了主要组织相容性复合体杂合性缺失(LOH)、等位基因特异性突变以及表达和抑制测量(MHC Hammer)。我们在正常肺部和乳腺组织中发现了 HLA 等位基因表达的广泛变异性和普遍存在的 HLA 替代剪接。在肺TRACERx和肺及乳腺TCGA队列中,61%的肺腺癌(LUAD)、76%的肺鳞癌(LUSC)和35%的雌激素受体阳性(ER+)癌症存在I类HLA转录抑制,而31%、11%和15%的LUAD、LUSC和ER+癌症存在HLA肿瘤丰富替代剪接。与HLA功能障碍在肿瘤演化中的重要性相一致的是,在LUAD中,HLA LOH与转移相关,而播种转移的LUAD原发肿瘤区域的有效新抗原负荷低于非播种区域。这些数据凸显了HLA转录组破坏的程度和重要性,包括抑制和替代剪接在癌症进化中的作用。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature genetics
生物-遗传学
CiteScore
43.00
自引率
2.60%
发文量
241
审稿时长
3 months
期刊介绍:
Nature Genetics publishes the very highest quality research in genetics. It encompasses genetic and functional genomic studies on human and plant traits and on other model organisms. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation.
Integrative genetic topics comprise, but are not limited to:
-Genes in the pathology of human disease
-Molecular analysis of simple and complex genetic traits
-Cancer genetics
-Agricultural genomics
-Developmental genetics
-Regulatory variation in gene expression
-Strategies and technologies for extracting function from genomic data
-Pharmacological genomics
-Genome evolution
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


