Rise and SINE: roles of transcription factors and retrotransposons in zygotic genome activation
IF 81.3
1区 生物学
Q1 CELL BIOLOGY
引用次数: 0
Abstract
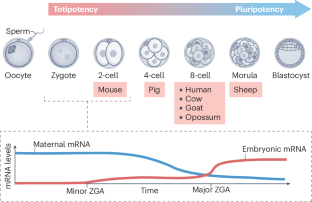
In sexually reproducing organisms, life begins with the fusion of transcriptionally silent gametes, the oocyte and sperm. Although initiation of transcription in the embryo, known as zygotic genome activation (ZGA), is universally required for development, the transcription factors regulating this process are poorly conserved. In this Perspective, we discuss recent insights into the mechanisms of ZGA in totipotent mammalian embryos, namely ZGA regulation by several transcription factors, including by orphan nuclear receptors (OrphNRs) such as the pioneer transcription factor NR5A2, and by factors of the DUX, TPRX and OBOX families. We performed a meta-analysis and compiled a list of pan-ZGA genes, and found that most of these genes are indeed targets of the above transcription factors. Remarkably, more than a third of these ZGA genes appear to be regulated both by OrphNRs such as NR5A2 and by OBOX proteins, whose motifs co-occur in SINE B1 retrotransposable elements, which are enriched near ZGA genes. We propose that ZGA in mice is activated by recruitment of multiple transcription factors to SINE B1 elements that function as enhancers, and discuss a potential relevance of this mechanism to Alu retrotransposable elements in human ZGA. Although zygotic genome activation (ZGA) is universally required for development, the responsible transcription factors are poorly conserved. In mammalian totipotent embryos, (pioneer) transcription factors of two families co-regulate many ZGA genes by binding to nearby SINE retrotransposons, which thus function as their enhancers.

崛起与SINE:转录因子和逆转录转座子在子代基因组激活中的作用
在有性生殖的生物体中,生命始于转录沉默配子(卵细胞和精子)的融合。虽然胚胎中转录的启动(称为子代基因组激活(ZGA))是发育的普遍需要,但调控这一过程的转录因子的保守性很差。在本《视角》中,我们讨论了最近对全能哺乳动物胚胎中 ZGA 机制的见解,即 ZGA 受多种转录因子调控,包括孤儿核受体(OrphNRs)(如先驱转录因子 NR5A2)以及 DUX、TPRX 和 OBOX 家族因子。我们进行了一项荟萃分析,编制了一份泛 ZGA 基因列表,发现其中大多数基因确实是上述转录因子的靶标。值得注意的是,在这些 ZGA 基因中,有三分之一以上的基因似乎同时受到 OrphNRs(如 NR5A2)和 OBOX 蛋白的调控,OBOX 蛋白的基序共同出现在 SINE B1 可逆转录元件中,而 SINE B1 可逆转录元件富集在 ZGA 基因附近。我们提出,小鼠的 ZGA 是通过将多种转录因子招募到起增强子作用的 SINE B1 元件而激活的,并讨论了这一机制与人类 ZGA 中的 Alu 可逆转录元件的潜在相关性。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
CiteScore
173.60
自引率
0.50%
发文量
118
审稿时长
6-12 weeks
期刊介绍:
Nature Reviews Molecular Cell Biology is a prestigious journal that aims to be the primary source of reviews and commentaries for the scientific communities it serves. The journal strives to publish articles that are authoritative, accessible, and enriched with easily understandable figures, tables, and other display items. The goal is to provide an unparalleled service to authors, referees, and readers, and the journal works diligently to maximize the usefulness and impact of each article. Nature Reviews Molecular Cell Biology publishes a variety of article types, including Reviews, Perspectives, Comments, and Research Highlights, all of which are relevant to molecular and cell biologists. The journal's broad scope ensures that the articles it publishes reach the widest possible audience.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


