From computational models of the splicing code to regulatory mechanisms and therapeutic implications
IF 39.1
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
Abstract
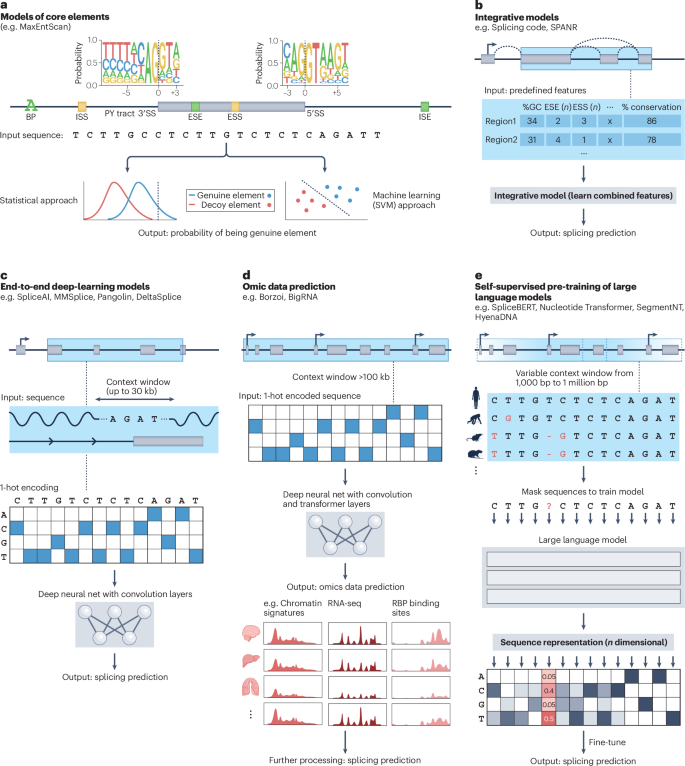
Since the discovery of RNA splicing and its role in gene expression, researchers have sought a set of rules, an algorithm or a computational model that could predict the splice isoforms, and their frequencies, produced from any transcribed gene in a specific cellular context. Over the past 30 years, these models have evolved from simple position weight matrices to deep-learning models capable of integrating sequence data across vast genomic distances. Most recently, new model architectures are moving the field closer to context-specific alternative splicing predictions, and advances in sequencing technologies are expanding the type of data that can be used to inform and interpret such models. Together, these developments are driving improved understanding of splicing regulatory mechanisms and emerging applications of the splicing code to the rational design of RNA- and splicing-based therapeutics. This Review describes how increasingly sophisticated omics data and computational models of the splicing code are paving the way to more accurate, context-specific splicing predictions, while also providing insights into the regulatory mechanisms and therapeutic applications of alternative splicing.

从剪接代码的计算模型到调控机制和治疗意义
自从发现 RNA 剪接及其在基因表达中的作用以来,研究人员一直在寻找一套规则、算法或计算模型,以预测在特定细胞环境中任何转录基因产生的剪接异构体及其频率。在过去的 30 年中,这些模型已经从简单的位置权重矩阵发展到能够整合跨巨大基因组距离的序列数据的深度学习模型。最近,新的模型架构使该领域更接近于特定背景下的替代剪接预测,而测序技术的进步则扩大了可用于告知和解释此类模型的数据类型。这些发展共同推动了人们对剪接调控机制的进一步了解,以及剪接密码在合理设计基于 RNA 和剪接的治疗方法方面的新兴应用。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Nature Reviews Genetics
生物-遗传学
CiteScore
57.40
自引率
0.50%
发文量
113
审稿时长
6-12 weeks
期刊介绍:
At Nature Reviews Genetics, our goal is to be the leading source of reviews and commentaries for the scientific communities we serve. We are dedicated to publishing authoritative articles that are easily accessible to our readers. We believe in enhancing our articles with clear and understandable figures, tables, and other display items. Our aim is to provide an unparalleled service to authors, referees, and readers, and we are committed to maximizing the usefulness and impact of each article we publish.
Within our journal, we publish a range of content including Research Highlights, Comments, Reviews, and Perspectives that are relevant to geneticists and genomicists. With our broad scope, we ensure that the articles we publish reach the widest possible audience.
As part of the Nature Reviews portfolio of journals, we strive to uphold the high standards and reputation associated with this esteemed collection of publications.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


