Proximity ligation-triggered DNAzyme for selective fluorescent aptasensing of methicillin-resistant Staphylococcus aureus
IF 4.6
Q2 MATERIALS SCIENCE, BIOMATERIALS
引用次数: 0
Abstract
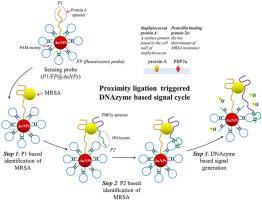
There is an urgent need for novel strategies to accurately and reliably detect pathogenic bacteria to address the global epidemic of antibiotic resistance. This study proposes an innovative approach combining dual aptamer-based target recognition and proximity ligation assay (PLA) triggered DNAzyme recycling cleavage. This method allows for the precise identification and reliable detection of methicillin-resistant Staphylococcus aureus (MRSA). The fluorescence probe labeled with a fluorophore is modified on gold nanoparticles (AuNPs), resulting in the quenching of the fluorescent signal by the AuNPs. The interaction between MRSA and two aptamers leads to forming a Mg2+-dependent DNAzyme. The DNAzyme cleaves the fluorescence probe, causing the fluorescent fragment to detach from the surface of the AuNPs, in which the quenched fluorescence signal in the fluorescence probe reappears. The DNAzyme-assisted cleavage and rebinding process generates a processive strolling along the surface track of AuNPs. Consequently, the fluorescence intensity experiences a substantial recovery. A strong linear correlation is observed between the fluorescence intensity and MRSA concentration within 50 cfu/mL to 106 cfu/mL. We believe that implementing the novel integrated strategy will broaden the range of bacterial detection methods in the battlefield environment and stimulate the creation of potential new drugs in the future.
用于耐甲氧西林金黄色葡萄球菌选择性荧光诱导的近接触发 DNA 酶。
为应对抗生素耐药性在全球的流行,我们迫切需要采用新的策略来准确可靠地检测病原菌。本研究提出了一种创新方法,它结合了基于双适配体的目标识别和近接检测(PLA)触发的 DNA 酶循环裂解。该方法可精确识别和可靠检测耐甲氧西林金黄色葡萄球菌(MRSA)。用荧光团标记的荧光探针被修饰在金纳米粒子(AuNPs)上,导致荧光信号被 AuNPs淬灭。MRSA 与两种适配体之间的相互作用会形成一种 Mg2+ 依赖性 DNA 酶。DNA 酶裂解荧光探针,使荧光片段从 AuNPs 表面脱离,荧光探针中被淬灭的荧光信号重新出现。DNA 酶辅助的裂解和重新结合过程产生了沿着 AuNPs 表面轨迹漫步的过程。因此,荧光强度出现了大幅恢复。在 50 cfu/mL 至 106 cfu/mL 的范围内,荧光强度与 MRSA 浓度之间存在很强的线性相关。我们相信,实施这种新型综合策略将拓宽战场环境中细菌检测方法的范围,并促进未来潜在新药的开发。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


