Directed evolution of a highly efficient TNA polymerase achieved by homologous recombination
IF 42.8
1区 化学
Q1 CHEMISTRY, PHYSICAL
引用次数: 0
Abstract
Reprogramming DNA polymerases to synthesize xeno-nucleic acids (XNAs) is an important challenge that tests current enzyme engineering tools. Here we describe an evolutionary campaign aimed at generating an XNA polymerase that can efficiently make α-l-threofuranosyl nucleic acid (TNA)—an artificial genetic polymer that is recalcitrant to nucleases and resistant to acid-mediated degradation. Starting from a homologous recombination library, iterative cycles of selection were performed to traverse the fitness landscape in search of neutral mutations with increased evolutionary potential. Subsequent directed evolution of focused mutagenic libraries yielded 10–92, a newly engineered TNA polymerase that functions with a catalytic rate of ∼1 nt s−1 and >99% fidelity. A crystal structure of the closed ternary complex reveals the degree of structural change required to remodel the active site pocket for improved TNA synthesis activity. Together, these data demonstrate the importance of recombination as a strategy for evolving XNA polymerases with considerable practical value for biotechnology and medicine. The catalytic power of DNA polymerases for artificial genetic polymer (XNA) synthesis remains underdeveloped. Now, the evolution and structure of an α-l-threofuranosyl nucleic acid polymerase is described that achieves XNA synthesis with ∼1 nt s−1 and >99% template-copying fidelity.
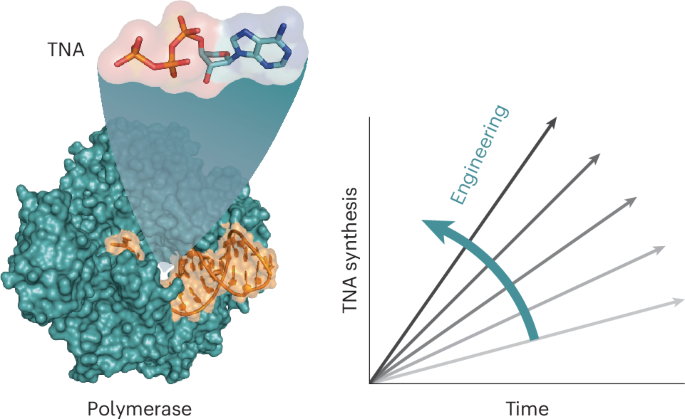
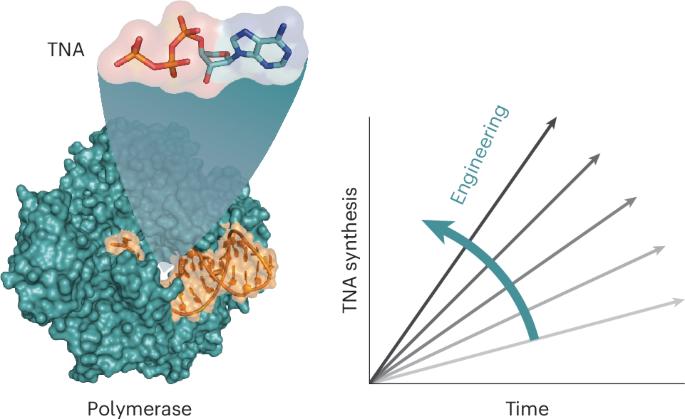
通过同源重组实现高效 TNA 聚合酶的定向进化
重新编程 DNA 聚合酶以合成异种核酸(XNA)是一项重要挑战,对现有的酶工程工具提出了考验。在这里,我们描述了一项旨在产生一种XNA聚合酶的进化活动,这种聚合酶可以有效地制造α-l-苏呋糖核酸(TNA)--一种对核酸酶有抵抗力、对酸介导的降解有抵抗力的人工基因聚合物。从同源重组文库开始,迭代选择循环遍历适合度景观,寻找具有更大进化潜力的中性突变。随后的定向进化聚焦诱变文库产生了 10-92,这是一种新设计的 TNA 聚合酶,其催化速率为 1 nt s-1 及 99% 的保真度。封闭三元复合物的晶体结构揭示了重塑活性位点口袋以提高 TNA 合成活性所需的结构变化程度。这些数据共同证明了重组作为 XNA 聚合酶进化策略的重要性,对生物技术和医学具有相当大的实用价值。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Nature Catalysis
Chemical Engineering-Bioengineering
CiteScore
52.10
自引率
1.10%
发文量
140
期刊介绍:
Nature Catalysis serves as a platform for researchers across chemistry and related fields, focusing on homogeneous catalysis, heterogeneous catalysis, and biocatalysts, encompassing both fundamental and applied studies. With a particular emphasis on advancing sustainable industries and processes, the journal provides comprehensive coverage of catalysis research, appealing to scientists, engineers, and researchers in academia and industry.
Maintaining the high standards of the Nature brand, Nature Catalysis boasts a dedicated team of professional editors, rigorous peer-review processes, and swift publication times, ensuring editorial independence and quality. The journal publishes work spanning heterogeneous catalysis, homogeneous catalysis, and biocatalysis, covering areas such as catalytic synthesis, mechanisms, characterization, computational studies, nanoparticle catalysis, electrocatalysis, photocatalysis, environmental catalysis, asymmetric catalysis, and various forms of organocatalysis.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


