Advancing DNA-based quantification of Pacific oyster larvae using a HTqPCR multi-marker approach
IF 1.8
3区 生物学
Q3 ECOLOGY
Journal of Experimental Marine Biology and Ecology
Pub Date : 2024-09-23
DOI:10.1016/j.jembe.2024.152055
引用次数: 0
Abstract
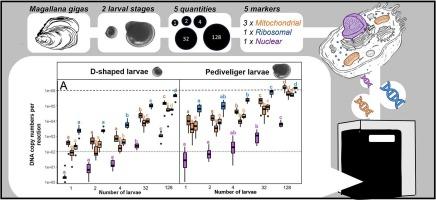
Marine bivalves play important roles in seafood supply/security and ecosystems services, with many aquaculture practices relying on wild stocks as sources of larvae. However, rapid and effective monitoring of larvae occurrence, abundance and distribution in the marine environment can be challenging. DNA-based approaches have shown promising results in providing novel monitoring tools. However, the effect of varying sources of different DNA targets and differential representation at distinct larval stages on genetic signal (copy numbers) are poorly understood. The present study used a High-Throughout real-time quantitative PCR (HTqPCR) platform to screen multiple genetic markers (originating from nuclear, ribosomal and mitochondrial genes) in DNA extracts from known number of larvae at two developmental stages (5-day old D-shape and 20-day old pediveliger), using the Pacific oysters (Magallana gigas) as a model study organism. Findings showed clear trends across sources of DNA, with ribosomal markers showing significantly higher numbers of DNA copies compared to mitochondrial markers, which in turn were significantly higher than nuclear markers. A predictable relationship was found between DNA copies found in five-day old D-shape larvae and 20-day old pediveliger larvae, indicating that developmental stages have significant effects on biomass estimation using DNA-derived data. Nuclear and mitochondrial markers showed higher accuracy for estimating higher amounts of larvae, whereas ribosomal markers showed higher accuracy for estimating lower numbers of larvae, suggesting that a multi-marker approach may be most appropriate. While this study provides empirical evidence on the effect of larval stages and source of DNA in quantifying bivalve larvae, it also provides a framework (enabled by a HTqPCR platform) to assess the suitability of DNA-based approaches for the monitoring of marine larvae occurrence as well as biomass.
利用 HTqPCR 多标记方法推进基于 DNA 的太平洋牡蛎幼体定量工作
海洋双壳贝类在海产品供应/安全和生态系统服务方面发挥着重要作用,许多水产养殖业都依赖野生种群作为幼体来源。然而,要快速有效地监测海洋环境中幼体的出现、数量和分布情况是一项挑战。基于 DNA 的方法在提供新型监测工具方面取得了可喜的成果。然而,人们对不同 DNA 目标的不同来源以及不同幼虫阶段的不同代表性对遗传信号(拷贝数)的影响知之甚少。本研究使用高通量实时定量 PCR(HTqPCR)平台,以太平洋牡蛎(Magallana gigas)为模型研究生物,在两个发育阶段(5 天大的 D 形幼体和 20 天大的足柄幼体)从已知数量的幼体 DNA 提取物中筛选多种遗传标记(来自核、核糖体和线粒体基因)。研究结果表明,不同来源的 DNA 有明显的变化趋势,核糖体标记的 DNA 拷贝数明显高于线粒体标记,而线粒体标记的 DNA 拷贝数又明显高于核标记。在 5 天大的 D 型幼虫和 20 天大的足鳞幼虫中发现的 DNA 拷贝数之间存在可预测的关系,这表明发育阶段对使用 DNA 衍生数据进行生物量估算有重大影响。核标记和线粒体标记在估算较多幼虫数量时显示出更高的准确性,而核糖体标记在估算较少幼虫数量时显示出更高的准确性,这表明多标记方法可能是最合适的。这项研究为量化双壳类幼体时幼体阶段和 DNA 来源的影响提供了经验证据,同时也提供了一个框架(由 HTqPCR 平台支持),用于评估基于 DNA 的方法是否适合监测海洋幼体的出现和生物量。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
CiteScore
4.30
自引率
0.00%
发文量
98
审稿时长
14 weeks
期刊介绍:
The Journal of Experimental Marine Biology and Ecology provides a forum for experimental ecological research on marine organisms in relation to their environment. Topic areas include studies that focus on biochemistry, physiology, behavior, genetics, and ecological theory. The main emphasis of the Journal lies in hypothesis driven experimental work, both from the laboratory and the field. Natural experiments or descriptive studies that elucidate fundamental ecological processes are welcome. Submissions should have a broad ecological framework beyond the specific study organism or geographic region.
Short communications that highlight emerging issues and exciting discoveries within five printed pages will receive a rapid turnaround. Papers describing important new analytical, computational, experimental and theoretical techniques and methods are encouraged and will be highlighted as Methodological Advances. We welcome proposals for Review Papers synthesizing a specific field within marine ecology. Finally, the journal aims to publish Special Issues at regular intervals synthesizing a particular field of marine science. All printed papers undergo a peer review process before being accepted and will receive a first decision within three months.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


