Systematic Evaluation of Affinity Enrichment Methods for O-GlcNAc Proteomics
IF 4.3
3区 材料科学
Q1 ENGINEERING, ELECTRICAL & ELECTRONIC
引用次数: 0
Abstract
O-Linked β-N-acetylglucosamine (O-GlcNAc) modification (i.e., O-GlcNAcylation) on proteins plays critical roles in the regulation of diverse biological processes. However, protein O-GlcNAcylation analysis, especially at a large scale, has been a challenge. So far, a number of enrichment materials and methods have been developed for site-specific O-GlcNAc proteomics in different biological settings. Despite the presence of multiple methods, their performance for the O-GlcNAc proteomics is largely unclear. In this work, by using the lysates of PANC-1 cells (a pancreatic cancer cell line), we provided a head-to-head comparison of three affinity enrichment methods and materials (i.e., antibody, lectin AANL6, and an OGA mutant) for site-specific O-GlcNAc proteomics. The enriched peptides were analyzed by HCD product-dependent EThcD (i.e., HCD-pd-EThcD) mass spectrometry. The resulting data files were processed by three different data analysis packages (i.e., Sequest HT, Byonic, and FragPipe). Our data suggest that each method captures a subpopulation of the O-GlcNAc proteins. Besides the enrichment methods, we also observe complementarity between the different data analysis tools. Thus, combining different approaches holds promise for enhanced coverage of O-GlcNAc proteomics.
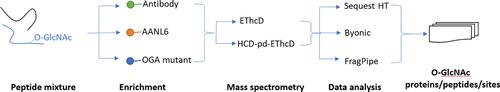
系统评估用于 O-谷氨酰核糖蛋白质组学的亲和富集方法
蛋白质上的 O-连接β-N-乙酰葡糖胺(O-GlcNAc)修饰(即 O-GlcNAcylation)在调控多种生物过程中发挥着关键作用。然而,蛋白质的 O-GlcNAcylation 分析,尤其是大规模分析,一直是个难题。迄今为止,已经开发出了许多富集材料和方法,用于在不同的生物环境中进行特定位点的 O-GlcNAc 蛋白组学研究。尽管存在多种方法,但它们在 O-GlcNAc 蛋白组学方面的性能还很不清楚。在这项工作中,我们利用 PANC-1 细胞(一种胰腺癌细胞系)的裂解液,对三种亲和富集方法和材料(即抗体、凝集素 AANL6 和 OGA 突变体)进行了正面比较,以进行特异性 O-GlcNAc 蛋白组学研究。富集的肽段通过 HCD 产物依赖性 EThcD(即 HCD-pd-EThcD)质谱进行分析。得到的数据文件由三种不同的数据分析软件包(即 Sequest HT、Byonic 和 FragPipe)处理。我们的数据表明,每种方法都能捕捉到 O-GlcNAc 蛋白的一个亚群。除了富集方法,我们还观察到不同数据分析工具之间的互补性。因此,结合不同的方法有望提高 O-GlcNAc 蛋白组学的覆盖率。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


