Synthetic macromolecular switches for precision control of therapeutic cell functions
IF 37.6
引用次数: 0
Abstract
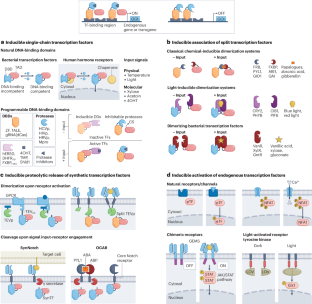
Cells rely on complex molecular networks to perceive and process external and internal signals into tailored responses. These cellular abilities can be augmented and modified by integrating artificial gene circuitry for the engineering of cell-based therapeutics. In this Review, we outline the engineering principles that govern the design of synthetic gene networks, highlighting how the sensitivity, detection range and specificity of synthetic gene networks can be optimized for in vivo functionality. In particular, we examine synthetic molecular modules, including transcriptionally regulated, translationally regulated and post-translationally regulated circuits, that enable tailored adjustments in therapeutic cell functions based on dynamic disease-state cues, or that can be remotely controlled using clinically compatible external molecular or physical signals. Furthermore, we explore the potential of multi-input regulatable logic-gated programs to enhance the efficacy and safety of engineered cell immunotherapies for cancer treatment, and highlight the application of synthetic gene circuits for gene therapy and the design of therapeutic microbes. Finally, we examine how synthetic-biology-inspired therapies may benefit from evolving genome engineering technologies and synergy with artificial intelligence. Synthetic gene circuits can endow cells with therapeutic functions. This Review discusses synthetic macromolecular systems, encompassing transcriptional, translational and post-translational mechanisms, that conditionally regulate protein expression and activity in response to specific internal or external stimuli, and their role in enhancing the therapeutic efficacy and safety of engineered cell therapies.

用于精确控制治疗细胞功能的合成大分子开关
细胞依靠复杂的分子网络来感知和处理外部和内部信号,并将其转化为量身定制的反应。这些细胞能力可以通过整合人工基因回路来增强和改变,从而实现基于细胞的治疗工程。在本综述中,我们将概述指导合成基因网络设计的工程原理,重点介绍如何优化合成基因网络的灵敏度、检测范围和特异性,以实现体内功能。特别是,我们研究了合成分子模块,包括转录调控、翻译调控和翻译后调控电路,这些模块可根据动态疾病状态线索对治疗细胞功能进行定制调整,或利用临床兼容的外部分子或物理信号进行远程控制。此外,我们还探讨了多输入可调节逻辑门控程序在提高工程细胞免疫疗法治疗癌症的有效性和安全性方面的潜力,并重点介绍了合成基因电路在基因治疗和治疗微生物设计方面的应用。最后,我们将探讨合成生物学启发疗法如何受益于不断发展的基因组工程技术以及与人工智能的协同作用。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


