Fasting shapes chromatin architecture through an mTOR/RNA Pol I axis
IF 17.3
1区 生物学
Q1 CELL BIOLOGY
引用次数: 0
Abstract
Chromatin architecture is a fundamental mediator of genome function. Fasting is a major environmental cue across the animal kingdom, yet how it impacts three-dimensional (3D) genome organization is unknown. Here we show that fasting induces an intestine-specific, reversible and large-scale spatial reorganization of chromatin in Caenorhabditis elegans. This fasting-induced 3D genome reorganization requires inhibition of the nutrient-sensing mTOR pathway, acting through the regulation of RNA Pol I, but not Pol II nor Pol III, and is accompanied by remodelling of the nucleolus. By uncoupling the 3D genome configuration from the animal’s nutritional status, we find that the expression of metabolic and stress-related genes increases when the spatial reorganization of chromatin occurs, showing that the 3D genome might support the transcriptional response in fasted animals. Our work documents a large-scale chromatin reorganization triggered by fasting and reveals that mTOR and RNA Pol I shape genome architecture in response to nutrients. Al-Refaie et al. show that fasting induces spatial reorganization of chromatin and formation of chromatin rings in an mTORC1- and RNA Pol I-dependent manner in the C. elegans intestine.
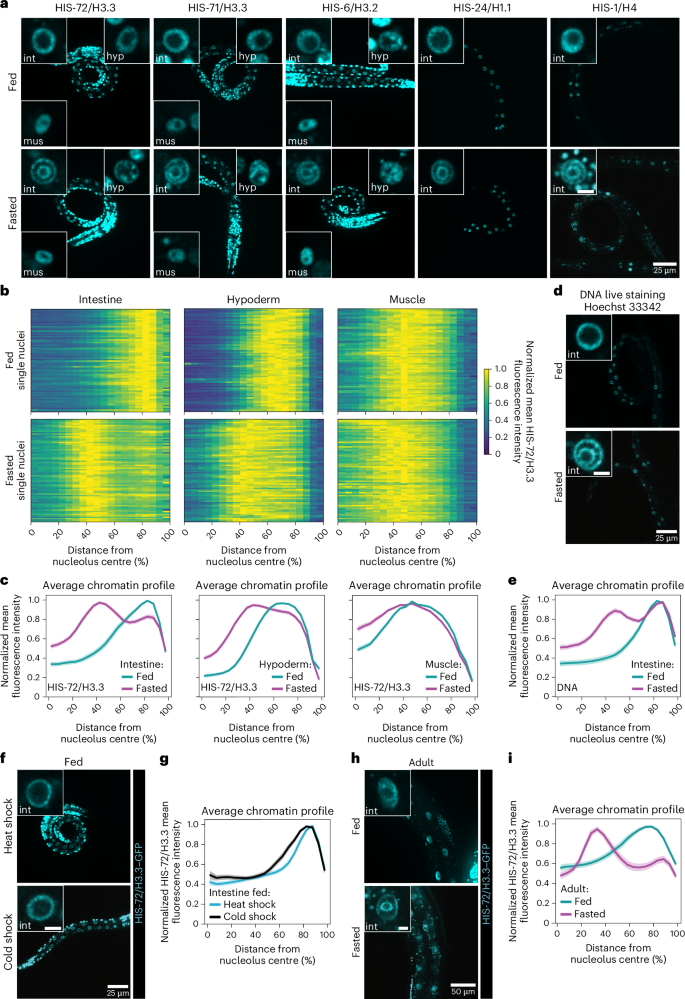
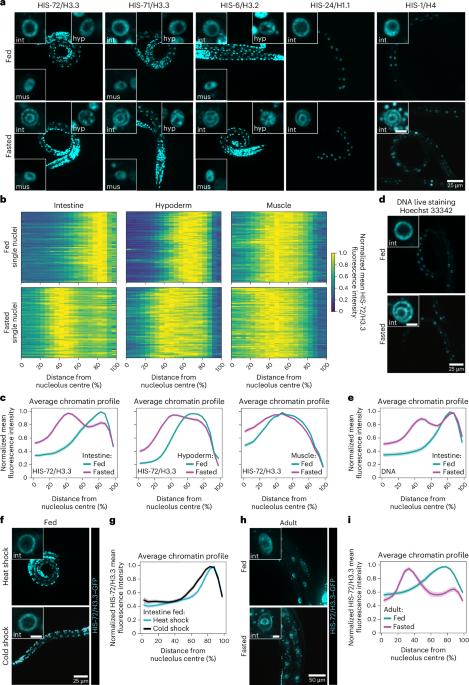
禁食通过 mTOR/RNA Pol I 轴塑造染色质结构
染色质结构是基因组功能的基本中介。禁食是整个动物界的一个主要环境线索,但它如何影响三维(3D)基因组的组织结构尚不清楚。在这里,我们发现禁食会诱导秀丽隐杆线虫肠道特异性、可逆性和大规模的染色质空间重组。这种禁食诱导的三维基因组重组需要通过调节 RNA Pol I(而不是 Pol II 或 Pol III)来抑制营养传感 mTOR 通路,并伴随着核仁的重塑。通过将三维基因组构型与动物的营养状况脱钩,我们发现当染色质发生空间重组时,代谢和应激相关基因的表达会增加,这表明三维基因组可能支持禁食动物的转录反应。我们的研究记录了由禁食引发的大规模染色质重组,并揭示了 mTOR 和 RNA Pol I 塑造基因组结构以应对营养物质。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Cell Biology
生物-细胞生物学
CiteScore
28.40
自引率
0.90%
发文量
219
审稿时长
3 months
期刊介绍:
Nature Cell Biology, a prestigious journal, upholds a commitment to publishing papers of the highest quality across all areas of cell biology, with a particular focus on elucidating mechanisms underlying fundamental cell biological processes. The journal's broad scope encompasses various areas of interest, including but not limited to:
-Autophagy
-Cancer biology
-Cell adhesion and migration
-Cell cycle and growth
-Cell death
-Chromatin and epigenetics
-Cytoskeletal dynamics
-Developmental biology
-DNA replication and repair
-Mechanisms of human disease
-Mechanobiology
-Membrane traffic and dynamics
-Metabolism
-Nuclear organization and dynamics
-Organelle biology
-Proteolysis and quality control
-RNA biology
-Signal transduction
-Stem cell biology
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


