Systematic prioritization of functional variants and effector genes underlying colorectal cancer risk
IF 31.7
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
Abstract
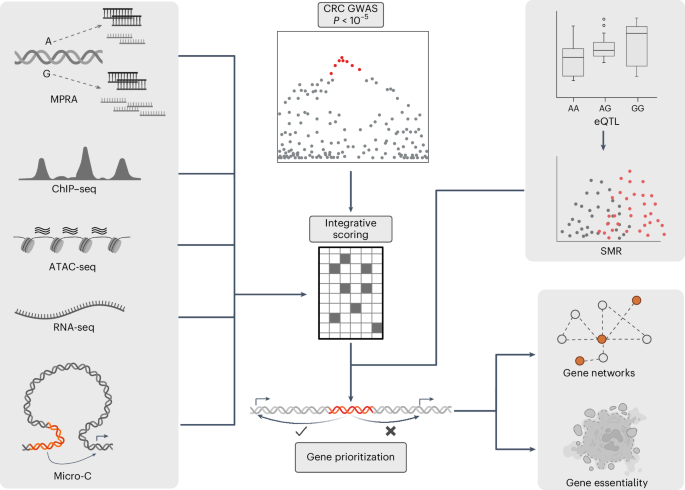
Genome-wide association studies of colorectal cancer (CRC) have identified 170 autosomal risk loci. However, for most of these, the functional variants and their target genes are unknown. Here, we perform statistical fine-mapping incorporating tissue-specific epigenetic annotations and massively parallel reporter assays to systematically prioritize functional variants for each CRC risk locus. We identify plausible causal variants for the 170 risk loci, with a single variant for 40. We link these variants to 208 target genes by analyzing colon-specific quantitative trait loci and implementing the activity-by-contact model, which integrates epigenomic features and Micro-C data, to predict enhancer–gene connections. By deciphering CRC risk loci, we identify direct links between risk variants and target genes, providing further insight into the molecular basis of CRC susceptibility and highlighting potential pharmaceutical targets for prevention and treatment. This study uses a combination of in silico and experimental techniques to ascribe target genes to 170 risk loci for colorectal cancer.
系统优先排序结直肠癌风险的功能变异和效应基因
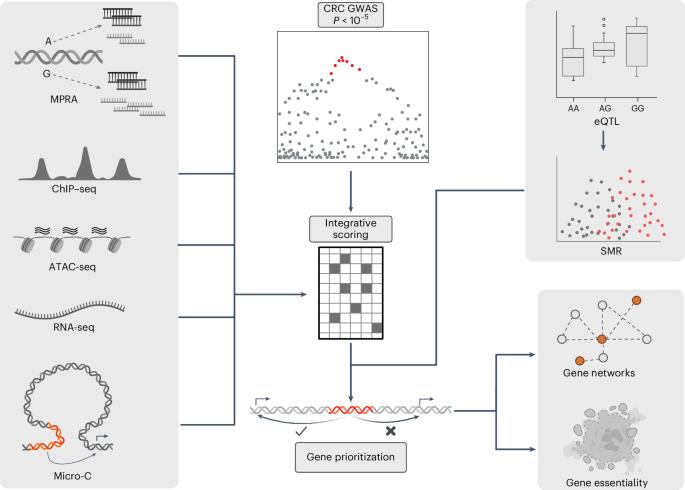
结直肠癌(CRC)的全基因组关联研究发现了 170 个常染色体风险位点。然而,其中大部分基因的功能变异及其靶基因尚不清楚。在这里,我们结合组织特异性表观遗传学注释和大规模并行报告基因检测进行了统计精细图谱绘制,以系统地确定每个 CRC 风险位点功能变异的优先次序。我们确定了 170 个风险基因座的似因变异,其中 40 个基因座只有一个变异。我们通过分析结肠特异性定量性状位点,将这些变异与 208 个目标基因联系起来,并实施了活性-接触模型(该模型整合了表观基因组特征和 Micro-C 数据)来预测增强子与基因的联系。通过破译 CRC 风险位点,我们确定了风险变异与目标基因之间的直接联系,从而进一步了解了 CRC 易感性的分子基础,并突出了预防和治疗的潜在药物靶点。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature genetics
生物-遗传学
CiteScore
43.00
自引率
2.60%
发文量
241
审稿时长
3 months
期刊介绍:
Nature Genetics publishes the very highest quality research in genetics. It encompasses genetic and functional genomic studies on human and plant traits and on other model organisms. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation.
Integrative genetic topics comprise, but are not limited to:
-Genes in the pathology of human disease
-Molecular analysis of simple and complex genetic traits
-Cancer genetics
-Agricultural genomics
-Developmental genetics
-Regulatory variation in gene expression
-Strategies and technologies for extracting function from genomic data
-Pharmacological genomics
-Genome evolution
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


