Mass Spectrometry-Based Top-Down Proteomics in Nanomedicine: Proteoform-Specific Measurement of Protein Corona
IF 15.8
1区 材料科学
Q1 CHEMISTRY, MULTIDISCIPLINARY
引用次数: 0
Abstract
Conventional mass spectrometry (MS)-based bottom-up proteomics (BUP) analysis of the protein corona [i.e., an evolving layer of biomolecules, mostly proteins, formed on the surface of nanoparticles (NPs) during their interactions with biomolecular fluids] enabled the nanomedicine community to partly identify the biological identity of NPs. Such an approach, however, fails to pinpoint the specific proteoforms─distinct molecular variants of proteins in the protein corona. The proteoform-level information could potentially advance the prediction of the biological fate and pharmacokinetics of nanomedicines. Recognizing this limitation, this study pioneers a robust and reproducible MS-based top-down proteomics (TDP) technique for characterizing proteoforms in the protein corona. Our TDP approach has successfully identified about 900 proteoforms in the protein corona of polystyrene NPs, ranging from 2 to 70 kDa, revealing proteoforms of 48 protein biomarkers with combinations of post-translational modifications, signal peptide cleavages, and/or truncations─details that BUP could not fully discern. This advancement in MS-based TDP offers a more advanced approach to characterize NP protein coronas, deepening our understanding of NPs’ biological identities. We, therefore, propose using both TDP and BUP strategies to obtain more comprehensive information about the protein corona, which, in turn, can further enhance the diagnostic and therapeutic efficacy of nanomedicine technologies.
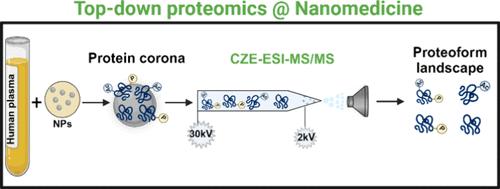
基于质谱的自上而下蛋白质组学在纳米医学中的应用:蛋白质形态特异性测量蛋白质电晕
传统的基于质谱(MS)的自下而上蛋白质组学(BUP)分析蛋白质电晕[即纳米粒子(NPs)与生物分子流体相互作用时在其表面形成的一层不断变化的生物大分子,主要是蛋白质]使纳米医学界能够部分识别 NPs 的生物特性。然而,这种方法无法确定特定的蛋白形态--蛋白质冠层中蛋白质的不同分子变体。蛋白质形态水平的信息有可能推动对纳米药物的生物学命运和药代动力学的预测。认识到这一局限性,本研究开创了一种稳健且可重复的基于质谱的自上而下蛋白质组学(TDP)技术,用于表征蛋白质电晕中的蛋白质形态。我们的 TDP 方法成功鉴定了聚苯乙烯 NP 蛋白电晕中约 900 种蛋白质形式,其范围从 2 kDa 到 70 kDa 不等,揭示了 48 种蛋白质生物标记物的蛋白质形式,这些蛋白质形式具有翻译后修饰、信号肽裂解和/或截短的组合--这些细节是 BUP 无法完全辨别的。基于 MS 的 TDP 的这一进步为表征 NP 蛋白冠层提供了更先进的方法,加深了我们对 NP 生物特性的了解。因此,我们建议同时使用 TDP 和 BUP 策略来获得更全面的蛋白质冠层信息,从而进一步提高纳米医学技术的诊断和治疗效果。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

ACS Nano
工程技术-材料科学:综合
CiteScore
26.00
自引率
4.10%
发文量
1627
审稿时长
1.7 months
期刊介绍:
ACS Nano, published monthly, serves as an international forum for comprehensive articles on nanoscience and nanotechnology research at the intersections of chemistry, biology, materials science, physics, and engineering. The journal fosters communication among scientists in these communities, facilitating collaboration, new research opportunities, and advancements through discoveries. ACS Nano covers synthesis, assembly, characterization, theory, and simulation of nanostructures, nanobiotechnology, nanofabrication, methods and tools for nanoscience and nanotechnology, and self- and directed-assembly. Alongside original research articles, it offers thorough reviews, perspectives on cutting-edge research, and discussions envisioning the future of nanoscience and nanotechnology.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


